Tutorial 2: Bayesian inference and decisions with continuous hidden state
Contents
Tutorial Objectives¶
Estimated timing of tutorial: 1 hour, 35 minutes
This notebook introduces you to Gaussians and Bayes’ rule for continuous distributions, allowing us to model simple put powerful combinations of prior information and new measurements. In this notebook you will work through the same ideas we explored in the binary state tutorial, but you will be introduced to a new problem: finding and guiding Astrocat! You will see this problem again in more complex ways in the following days.
In this notebook, you will:
Learn about the Gaussian distribution and its nice properties
Explore how we can extend the ideas from the binary hidden tutorial to continuous distributions
Explore how different priors can produce more complex posteriors.
Explore Loss functions often used in inference and complex utility functions.
Setup¶
⚠ Experimental LLM-enhanced tutorial ⚠
This notebook includes Neuromatch’s experimental Chatify 🤖 functionality. The Chatify notebook extension adds support for a large language model-based “coding tutor” to the materials. The tutor provides automatically generated text to help explain any code cell in this notebook.
Note that using Chatify may cause breaking changes and/or provide incorrect or misleading information. If you wish to proceed by installing and enabling the Chatify extension, you should run the next two code blocks (hidden by default). If you do not want to use this experimental version of the Neuromatch materials, please use the stable materials instead.
To use the Chatify helper, insert the %%explain magic command at the start of any code cell and then run it (shift + enter) to access an interface for receiving LLM-based assitance. You can then select different options from the dropdown menus depending on what sort of assitance you want. To disable Chatify and run the code block as usual, simply delete the %%explain command and re-run the cell.
Note that, by default, all of Chatify’s responses are generated locally. This often takes several minutes per response. Once you click the “Submit request” button, just be patient– stuff is happening even if you can’t see it right away!
Thanks for giving Chatify a try! Love it? Hate it? Either way, we’d love to hear from you about your Chatify experience! Please consider filling out our brief survey to provide feedback and help us make Chatify more awesome!
Run the next two cells to install and configure Chatify…
%pip install -q davos
import davos
davos.config.suppress_stdout = True
Note: you may need to restart the kernel to use updated packages.
smuggle chatify # pip: git+https://github.com/ContextLab/chatify.git
%load_ext chatify
Downloading and initializing model; this may take a few minutes...
llama.cpp: loading model from /home/runner/.cache/huggingface/hub/models--TheBloke--Llama-2-7B-Chat-GGML/snapshots/501a3c8182cd256a859888fff4e838c049d5d7f6/llama-2-7b-chat.ggmlv3.q5_1.bin
llama_model_load_internal: format = ggjt v3 (latest)
llama_model_load_internal: n_vocab = 32000
llama_model_load_internal: n_ctx = 512
llama_model_load_internal: n_embd = 4096
llama_model_load_internal: n_mult = 256
llama_model_load_internal: n_head = 32
llama_model_load_internal: n_layer = 32
llama_model_load_internal: n_rot = 128
llama_model_load_internal: freq_base = 10000.0
llama_model_load_internal: freq_scale = 1
llama_model_load_internal: ftype = 9 (mostly Q5_1)
llama_model_load_internal: n_ff = 11008
llama_model_load_internal: model size = 7B
llama_model_load_internal: ggml ctx size = 0.08 MB
llama_model_load_internal: mem required = 6390.60 MB (+ 1026.00 MB per state)
llama_new_context_with_model: kv self size = 256.00 MB
AVX = 1 | AVX2 = 1 | AVX512 = 1 | AVX512_VBMI = 0 | AVX512_VNNI = 0 | FMA = 1 | NEON = 0 | ARM_FMA = 0 | F16C = 1 | FP16_VA = 0 | WASM_SIMD = 0 | BLAS = 0 | SSE3 = 1 | VSX = 0 |
Install and import feedback gadget¶
# @title Install and import feedback gadget
!pip3 install vibecheck datatops --quiet
from vibecheck import DatatopsContentReviewContainer
def content_review(notebook_section: str):
return DatatopsContentReviewContainer(
"", # No text prompt
notebook_section,
{
"url": "https://pmyvdlilci.execute-api.us-east-1.amazonaws.com/klab",
"name": "neuromatch_cn",
"user_key": "y1x3mpx5",
},
).render()
feedback_prefix = "W3D1_T2"
# Imports
import numpy as np
import matplotlib.pyplot as plt
from scipy.stats import multivariate_normal
from scipy.stats import gamma as gamma_distribution
from matplotlib.transforms import Affine2D
Figure Settings¶
# @title Figure Settings
import logging
logging.getLogger('matplotlib.font_manager').disabled = True
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import ipywidgets as widgets
from IPython.display import clear_output
from ipywidgets import FloatSlider, Dropdown, interactive_output
from ipywidgets import interact, fixed, HBox, Layout, VBox, interactive, Label
plt.style.use("https://raw.githubusercontent.com/NeuromatchAcademy/course-content/main/nma.mplstyle")
import warnings
warnings.filterwarnings("ignore")
Plotting Functions (part 1)¶
# @title Plotting Functions (part 1)
def plot_mixture_prior(x, gaussian1, gaussian2, combined):
""" Plots a prior made of a mixture of gaussians
Args:
x (numpy array of floats): points at which the likelihood has been evaluated
gaussian1 (numpy array of floats): normalized probabilities for Gaussian 1 evaluated at each `x`
gaussian2 (numpy array of floats): normalized probabilities for Gaussian 2 evaluated at each `x`
posterior (numpy array of floats): normalized probabilities for the posterior evaluated at each `x`
Returns:
Nothing
"""
fig, ax = plt.subplots()
ax.plot(x, gaussian1, '--b', LineWidth=2, label='Gaussian 1')
ax.plot(x, gaussian2, '-.b', LineWidth=2, label='Gaussian 2')
ax.plot(x, combined, '-r', LineWidth=2, label='Gaussian Mixture')
ax.legend()
ax.set_ylabel('Probability')
ax.set_xlabel('Orientation (Degrees)')
def plot_gaussian(μ, σ):
x = np.linspace(-7, 7, 1000, endpoint=True)
y = gaussian(x, μ, σ)
plt.figure(figsize=(6, 4))
plt.plot(x, y, c='blue')
plt.fill_between(x, y, color='b', alpha=0.2)
plt.ylabel('$\mathcal{N}(x, \mu, \sigma^2)$')
plt.xlabel('x')
plt.yticks([])
plt.show()
def plot_losses(μ, σ):
x = np.linspace(-2, 2, 400, endpoint=True)
y = gaussian(x, μ, σ)
error = x - μ
mse_loss = (error)**2
abs_loss = np.abs(error)
zero_one_loss = (np.abs(error) >= 0.02).astype(np.float)
fig, (ax_gaus, ax_error) = plt.subplots(2, 1, figsize=(6, 8))
ax_gaus.plot(x, y, color='blue', label='true distribution')
ax_gaus.fill_between(x, y, color='blue', alpha=0.2)
ax_gaus.set_ylabel('$\\mathcal{N}(x, \\mu, \\sigma^2)$')
ax_gaus.set_xlabel('x')
ax_gaus.set_yticks([])
ax_gaus.legend(loc='upper right')
ax_error.plot(x, mse_loss, color='c', label='Mean Squared Error', linewidth=3)
ax_error.plot(x, abs_loss, color='m', label='Absolute Error', linewidth=3)
ax_error.plot(x, zero_one_loss, color='y', label='Zero-One Loss', linewidth=3)
ax_error.legend(loc='upper right')
ax_error.set_xlabel('$\\hat{\\mu}$')
ax_error.set_ylabel('Error')
plt.show()
def gaussian_mixture(mu1, mu2, sigma1, sigma2, factor):
assert 0.0 < factor < 1.0
x = np.linspace(-7.0, 7.0, 1000, endpoint=True)
y_1 = gaussian(x, mu1, sigma1)
y_2 = gaussian(x, mu2, sigma2)
mixture = y_1 * factor + y_2 * (1.0 - factor)
plt.figure(figsize=(8, 6))
plt.plot(x, y_1, c='deepskyblue', label='p(x)', linewidth=3.0)
plt.fill_between(x, y_1, color='deepskyblue', alpha=0.2)
plt.plot(x, y_2, c='aquamarine', label='q(x)', linewidth=3.0)
plt.fill_between(x, y_2, color='aquamarine', alpha=0.2)
plt.plot(x, mixture, c='b', label='$\pi \cdot p(x) + (1-\pi) \cdot q(x)$', linewidth=3.0)
plt.fill_between(x, mixture, color='b', alpha=0.2)
plt.yticks([])
plt.legend(loc="upper left")
# plt.ylabel('$f(x)$')
plt.xlabel('x')
plt.show()
def plot_utility_mixture_dist(mu1, sigma1, mu2, sigma2, mu_g, sigma_g,
mu_loc, mu_dist, plot_utility_row=True):
x = np.linspace(-7, 7, 1000, endpoint=True)
prior = gaussian(x, mu1, sigma1)
likelihood = gaussian(x, mu2, sigma2)
mu_post, sigma_post = product_guassian(mu1, mu2, sigma1, sigma2)
posterior = gaussian(x, mu_post, sigma_post)
gain = gaussian(x, mu_g, sigma_g)/2
sigma_mix, factor = 1.0, 0.5
mu_mix1, mu_mix2 = mu_loc - mu_dist/2, mu_loc + mu_dist/2
gaus_mix1, gaus_mix2 = gaussian(x, mu_mix1, sigma_mix), gaussian(x, mu_mix2, sigma_mix)
loss = factor * gaus_mix1 + (1 - factor) * gaus_mix2
utility = np.multiply(posterior, gain) - np.multiply(posterior, loss)
if plot_utility_row:
plot_bayes_utility_rows(x, prior, likelihood, posterior, gain, loss, utility)
else:
plot_bayes_row(x, prior, likelihood, posterior)
return None
def plot_mvn2d(mu1, mu2, sigma1, sigma2, corr):
x, y = np.mgrid[-2:2:.02, -2:2:.02]
cov12 = corr * sigma1 * sigma2
z = mvn2d(x, y, mu1, mu2, sigma1, sigma2, cov12)
plt.figure(figsize=(6, 6))
plt.contourf(x, y, z, cmap='Reds')
plt.axis("off")
plt.show()
def plot_marginal(sigma1, sigma2, c_x, c_y, corr):
mu1, mu2 = 0.0, 0.0
cov12 = corr * sigma1 * sigma2
xx, yy = np.mgrid[-2:2:.02, -2:2:.02]
x, y = xx[:, 0], yy[0]
p_x = gaussian(x, mu1, sigma1)
p_y = gaussian(y, mu2, sigma2)
zz = mvn2d(xx, yy, mu1, mu2, sigma1, sigma2, cov12)
mu_x_y = mu1+cov12*(c_y-mu2)/sigma2**2
mu_y_x = mu2+cov12*(c_x-mu1)/sigma1**2
sigma_x_y = np.sqrt(sigma1**2 - cov12**2/sigma2**2)
sigma_y_x = np.sqrt(sigma2**2 - cov12**2/sigma1**2)
p_x_y = gaussian(x, mu_x_y, sigma_x_y)
p_y_x = gaussian(x, mu_y_x, sigma_y_x)
p_c_y = gaussian(mu_x_y-sigma_x_y, mu_x_y, sigma_x_y)
p_c_x = gaussian(mu_y_x-sigma_y_x, mu_y_x, sigma_y_x)
# definitions for the axes
left, width = 0.1, 0.65
bottom, height = 0.1, 0.65
spacing = 0.01
rect_z = [left, bottom, width, height]
rect_x = [left, bottom + height + spacing, width, 0.2]
rect_y = [left + width + spacing, bottom, 0.2, height]
# start with a square Figure
fig = plt.figure(figsize=(8, 8))
ax_z = fig.add_axes(rect_z)
ax_x = fig.add_axes(rect_x, sharex=ax_z)
ax_y = fig.add_axes(rect_y, sharey=ax_z)
ax_z.set_axis_off()
ax_x.set_axis_off()
ax_y.set_axis_off()
ax_x.set_xlim(np.min(x), np.max(x))
ax_y.set_ylim(np.min(y), np.max(y))
ax_z.contourf(xx, yy, zz, cmap='Greys')
ax_z.hlines(c_y, mu_x_y-sigma_x_y, mu_x_y+sigma_x_y, color='c', zorder=9, linewidth=3)
ax_z.vlines(c_x, mu_y_x-sigma_y_x, mu_y_x+sigma_y_x, color='m', zorder=9, linewidth=3)
ax_x.plot(x, p_x, label='$p(x)$', c = 'b', linewidth=3)
ax_x.plot(x, p_x_y, label='$p(x|y = C_y)$', c = 'c', linestyle='dashed', linewidth=3)
ax_x.hlines(p_c_y, mu_x_y-sigma_x_y, mu_x_y+sigma_x_y, color='c', linestyle='dashed', linewidth=3)
ax_y.plot(p_y, y, label='$p(y)$', c = 'r', linewidth=3)
ax_y.plot(p_y_x, y, label='$p(y|x = C_x)$', c = 'm', linestyle='dashed', linewidth=3)
ax_y.vlines(p_c_x, mu_y_x-sigma_y_x, mu_y_x+sigma_y_x, color='m', linestyle='dashed', linewidth=3)
ax_x.legend(loc="upper left", frameon=False)
ax_y.legend(loc="lower right", frameon=False)
plt.show()
def plot_bayes(mu1, mu2, sigma1, sigma2):
x = np.linspace(-7, 7, 1000, endpoint=True)
prior = gaussian(x, mu1, sigma1)
likelihood = gaussian(x, mu2, sigma2)
mu_post, sigma_post = product_guassian(mu1, mu2, sigma1, sigma2)
posterior = gaussian(x, mu_post, sigma_post)
plt.figure(figsize=(8, 6))
plt.plot(x, prior, c='b', label='prior')
plt.fill_between(x, prior, color='b', alpha=0.2)
plt.plot(x, likelihood, c='r', label='likelihood')
plt.fill_between(x, likelihood, color='r', alpha=0.2)
plt.plot(x, posterior, c='k', label='posterior')
plt.fill_between(x, posterior, color='k', alpha=0.2)
plt.yticks([])
plt.legend(loc="upper left")
plt.ylabel('$\mathcal{N}(x, \mu, \sigma^2)$')
plt.xlabel('x')
plt.show()
def plot_information(mu1, sigma1, mu2, sigma2):
x = np.linspace(-7, 7, 1000, endpoint=True)
mu3, sigma3 = product_guassian(mu1, mu2, sigma1, sigma2)
prior = gaussian(x, mu1, sigma1)
likelihood = gaussian(x, mu2, sigma2)
posterior = gaussian(x, mu3, sigma3)
plt.figure(figsize=(8, 6))
plt.plot(x, prior, c='b', label='Satellite')
plt.fill_between(x, prior, color='b', alpha=0.2)
plt.plot(x, likelihood, c='r', label='Space Mouse')
plt.fill_between(x, likelihood, color='r', alpha=0.2)
plt.plot(x, posterior, c='k', label='Center')
plt.fill_between(x, posterior, color='k', alpha=0.2)
plt.yticks([])
plt.legend(loc="upper left")
plt.ylabel('$\mathcal{N}(x, \mu, \sigma^2)$')
plt.xlabel('x')
plt.show()
def plot_information_global(mu3, sigma3, mu1, mu2):
x = np.linspace(-7, 7, 1000, endpoint=True)
sigma1, sigma2 = reverse_product(mu3, sigma3, mu1, mu2)
prior = gaussian(x, mu1, sigma1)
likelihood = gaussian(x, mu2, sigma2)
posterior = gaussian(x, mu3, sigma3)
plt.figure(figsize=(8, 6))
plt.plot(x, prior, c='b', label='Satellite')
plt.fill_between(x, prior, color='b', alpha=0.2)
plt.plot(x, likelihood, c='r', label='Space Mouse')
plt.fill_between(x, likelihood, color='r', alpha=0.2)
plt.plot(x, posterior, c='k', label='Center')
plt.fill_between(x, posterior, color='k', alpha=0.2)
plt.yticks([])
plt.legend(loc="upper left")
plt.ylabel('$\mathcal{N}(x, \mu, \sigma^2)$')
plt.xlabel('x')
plt.show()
Plotting Functions (part 2)¶
# @title Plotting Functions (part 2)
def plot_loss_utility_gaussian(loss_f, mu, sigma, mu_true):
x = np.linspace(-7, 7, 1000, endpoint=True)
posterior = gaussian(x, mu, sigma)
y_label = "$p(x)$"
plot_loss_utility(x, posterior, loss_f, mu_true, y_label)
def plot_loss_utility_mixture(loss_f, mu1, mu2, sigma1, sigma2, factor, mu_true):
x = np.linspace(-7, 7, 1000, endpoint=True)
y_1 = gaussian(x, mu1, sigma1)
y_2 = gaussian(x, mu2, sigma2)
posterior = y_1 * factor + y_2 * (1.0 - factor)
y_label = "$\pi \cdot p(x) + (1-\pi) \cdot q(x)$"
plot_loss_utility(x, posterior, loss_f, mu_true, y_label)
def plot_loss_utility(x, posterior, loss_f, mu_true, y_label):
mean, median, mode = calc_mean_mode_median(x, posterior)
loss = calc_loss_func(loss_f, mu_true, x)
utility = calc_expected_loss(loss_f, posterior, x)
min_expected_loss = x[np.argmin(utility)]
plt.figure(figsize=(12, 8))
plt.subplot(2, 2, 1)
plt.title("Probability")
plt.plot(x, posterior, c='b')
plt.fill_between(x, posterior, color='b', alpha=0.2)
plt.yticks([])
plt.xlabel('x')
plt.ylabel(y_label)
plt.axvline(mean, ls='dashed', color='red', label='Mean')
plt.axvline(median, ls='dashdot', color='blue', label='Median')
plt.axvline(mode, ls='dotted', color='green', label='Mode')
plt.legend(loc="upper left")
plt.subplot(2, 2, 2)
plt.title(loss_f)
plt.plot(x, loss, c='c', label=loss_f)
# plt.fill_between(x, loss, color='c', alpha=0.2)
plt.ylabel('loss')
# plt.legend(loc="upper left")
plt.xlabel('x')
plt.subplot(2, 2, 3)
plt.title("Expected Loss")
plt.plot(x, utility, c='y', label='$\mathbb{E}[L]$')
plt.axvline(min_expected_loss, ls='dashed', color='red', label='$Min~ \mathbb{E}[Loss]$')
# plt.fill_between(x, utility, color='y', alpha=0.2)
plt.legend(loc="lower right")
plt.xlabel('x')
plt.ylabel('$\mathbb{E}[L]$')
plt.show()
def plot_loss_utility_bayes(mu1, mu2, sigma1, sigma2, mu_true, loss_f):
x = np.linspace(-4, 4, 1000, endpoint=True)
prior = gaussian(x, mu1, sigma1)
likelihood = gaussian(x, mu2, sigma2)
mu_post, sigma_post = product_guassian(mu1, mu2, sigma1, sigma2)
posterior = gaussian(x, mu_post, sigma_post)
loss = calc_loss_func(loss_f, mu_true, x)
utility = - calc_expected_loss(loss_f, posterior, x)
plt.figure(figsize=(18, 5))
plt.subplot(1, 3, 1)
plt.title("Posterior distribution")
plt.plot(x, prior, c='b', label='prior')
plt.fill_between(x, prior, color='b', alpha=0.2)
plt.plot(x, likelihood, c='r', label='likelihood')
plt.fill_between(x, likelihood, color='r', alpha=0.2)
plt.plot(x, posterior, c='k', label='posterior')
plt.fill_between(x, posterior, color='k', alpha=0.2)
plt.yticks([])
plt.legend(loc="upper left")
# plt.ylabel('$f(x)$')
plt.xlabel('x')
plt.subplot(1, 3, 2)
plt.title(loss_f)
plt.plot(x, loss, c='c')
# plt.fill_between(x, loss, color='c', alpha=0.2)
plt.ylabel('loss')
plt.subplot(1, 3, 3)
plt.title("Expected utility")
plt.plot(x, utility, c='y', label='utility')
# plt.fill_between(x, utility, color='y', alpha=0.2)
plt.legend(loc="upper left")
plt.show()
def plot_simple_utility_gaussian(mu, sigma, mu_g, mu_c, sigma_g, sigma_c):
x = np.linspace(-7, 7, 1000, endpoint=True)
posterior = gaussian(x, mu, sigma)
gain = gaussian(x, mu_g, sigma_g)
loss = gaussian(x, mu_c, sigma_c)
utility = np.multiply(posterior, gain) - np.multiply(posterior, loss)
plt.figure(figsize=(15, 5))
plt.subplot(1, 3, 1)
plt.title("Probability")
plt.plot(x, posterior, c='b', label='posterior')
plt.fill_between(x, posterior, color='b', alpha=0.2)
plt.yticks([])
# plt.legend(loc="upper left")
plt.xlabel('x')
plt.subplot(1, 3, 2)
plt.title("utility function")
plt.plot(x, gain, c='m', label='gain')
# plt.fill_between(x, gain, color='m', alpha=0.2)
plt.plot(x, -loss, c='c', label='loss')
# plt.fill_between(x, -loss, color='c', alpha=0.2)
plt.legend(loc="upper left")
plt.subplot(1, 3, 3)
plt.title("expected utility")
plt.plot(x, utility, c='y', label='utility')
# plt.fill_between(x, utility, color='y', alpha=0.2)
plt.legend(loc="upper left")
plt.show()
def plot_utility_gaussian(mu1, mu2, sigma1, sigma2, mu_g, mu_c, sigma_g, sigma_c, plot_utility_row=True):
x = np.linspace(-7, 7, 1000, endpoint=True)
prior = gaussian(x, mu1, sigma1)
likelihood = gaussian(x, mu2, sigma2)
mu_post, sigma_post = product_guassian(mu1, mu2, sigma1, sigma2)
posterior = gaussian(x, mu_post, sigma_post)
if plot_utility_row:
gain = gaussian(x, mu_g, sigma_g)
loss = gaussian(x, mu_c, sigma_c)
utility = np.multiply(posterior, gain) - np.multiply(posterior, loss)
plot_bayes_utility_rows(x, prior, likelihood, posterior, gain, loss, utility)
else:
plot_bayes_row(x, prior, likelihood, posterior)
return None
def plot_utility_mixture(mu_m1, mu_m2, sigma_m1, sigma_m2, factor,
mu, sigma, mu_g, mu_c, sigma_g, sigma_c, plot_utility_row=True):
x = np.linspace(-7, 7, 1000, endpoint=True)
y_1 = gaussian(x, mu_m1, sigma_m1)
y_2 = gaussian(x, mu_m2, sigma_m2)
prior = y_1 * factor + y_2 * (1.0 - factor)
likelihood = gaussian(x, mu, sigma)
posterior = np.multiply(prior, likelihood)
posterior = posterior / (posterior.sum() * (x[1] - x[0]))
if plot_utility_row:
gain = gaussian(x, mu_g, sigma_g)
loss = gaussian(x, mu_c, sigma_c)
utility = np.multiply(posterior, gain) - np.multiply(posterior, loss)
plot_bayes_utility_rows(x, prior, likelihood, posterior, gain, loss, utility)
else:
plot_bayes_row(x, prior, likelihood, posterior)
return None
def plot_utility_uniform(mu, sigma, mu_g, mu_c, sigma_g, sigma_c, plot_utility_row=True):
x = np.linspace(-7, 7, 1000, endpoint=True)
prior = np.ones_like(x) / (x.max() - x.min())
likelihood = gaussian(x, mu, sigma)
posterior = likelihood
# posterior = np.multiply(prior, likelihood)
# posterior = posterior / (posterior.sum() * (x[1] - x[0]))
if plot_utility_row:
gain = gaussian(x, mu_g, sigma_g)
loss = gaussian(x, mu_c, sigma_c)
utility = np.multiply(posterior, gain) - np.multiply(posterior, loss)
plot_bayes_utility_rows(x, prior, likelihood, posterior, gain, loss, utility)
else:
plot_bayes_row(x, prior, likelihood, posterior)
return None
def plot_utility_gamma(alpha, beta, offset, mu, sigma, mu_g, mu_c, sigma_g, sigma_c, plot_utility_row=True):
x = np.linspace(-4, 10, 1000, endpoint=True)
prior = gamma_pdf(x-offset, alpha, beta)
likelihood = gaussian(x, mu, sigma)
posterior = np.multiply(prior, likelihood)
posterior = posterior / (posterior.sum() * (x[1] - x[0]))
if plot_utility_row:
gain = gaussian(x, mu_g, sigma_g)
loss = gaussian(x, mu_c, sigma_c)
utility = np.multiply(posterior, gain) - np.multiply(posterior, loss)
plot_bayes_utility_rows(x, prior, likelihood, posterior, gain, loss, utility)
else:
plot_bayes_row(x, prior, likelihood, posterior)
return None
def plot_bayes_row(x, prior, likelihood, posterior):
mean, median, mode = calc_mean_mode_median(x, posterior)
plt.figure(figsize=(12, 4))
plt.subplot(1, 2, 1)
plt.title("Prior and likelihood distribution")
plt.plot(x, prior, c='b', label='prior')
plt.fill_between(x, prior, color='b', alpha=0.2)
plt.plot(x, likelihood, c='r', label='likelihood')
plt.fill_between(x, likelihood, color='r', alpha=0.2)
# plt.plot(x, posterior, c='k', label='posterior')
# plt.fill_between(x, posterior, color='k', alpha=0.2)
plt.yticks([])
plt.legend(loc="upper left")
# plt.ylabel('$f(x)$')
plt.xlabel('x')
plt.subplot(1, 2, 2)
plt.title("Posterior distribution")
plt.plot(x, posterior, c='k', label='posterior')
plt.fill_between(x, posterior, color='k', alpha=0.1)
plt.axvline(mean, ls='dashed', color='red', label='Mean')
plt.axvline(median, ls='dashdot', color='blue', label='Median')
plt.axvline(mode, ls='dotted', color='green', label='Mode')
plt.legend(loc="upper left")
plt.yticks([])
plt.xlabel('x')
plt.show()
def plot_bayes_utility_rows(x, prior, likelihood, posterior, gain, loss, utility):
mean, median, mode = calc_mean_mode_median(x, posterior)
max_utility = x[np.argmax(utility)]
plt.figure(figsize=(12, 8))
plt.subplot(2, 2, 1)
plt.title("Prior and likelihood distribution")
plt.plot(x, prior, c='b', label='prior')
plt.fill_between(x, prior, color='b', alpha=0.2)
plt.plot(x, likelihood, c='r', label='likelihood')
plt.fill_between(x, likelihood, color='r', alpha=0.2)
# plt.plot(x, posterior, c='k', label='posterior')
# plt.fill_between(x, posterior, color='k', alpha=0.2)
plt.yticks([])
plt.legend(loc="upper left")
# plt.ylabel('$f(x)$')
plt.xlabel('x')
plt.subplot(2, 2, 2)
plt.title("Posterior distribution")
plt.plot(x, posterior, c='k', label='posterior')
plt.fill_between(x, posterior, color='k', alpha=0.1)
plt.axvline(mean, ls='dashed', color='red', label='Mean')
plt.axvline(median, ls='dashdot', color='blue', label='Median')
plt.axvline(mode, ls='dotted', color='green', label='Mode')
plt.legend(loc="upper left")
plt.yticks([])
plt.xlabel('x')
plt.subplot(2, 2, 3)
plt.title("utility function")
plt.plot(x, gain, c='m', label='gain')
# plt.fill_between(x, gain, color='m', alpha=0.2)
plt.plot(x, -loss, c='c', label='loss')
# plt.fill_between(x, -loss, color='c', alpha=0.2)
plt.legend(loc="upper left")
plt.xlabel('x')
plt.subplot(2, 2, 4)
plt.title("expected utility")
plt.plot(x, utility, c='y', label='utility')
# plt.fill_between(x, utility, color='y', alpha=0.2)
plt.axvline(max_utility, ls='dashed', color='red', label='Max utility')
plt.legend(loc="upper left")
plt.xlabel('x')
plt.ylabel('utility')
plt.legend(loc="lower right")
plt.show()
def plot_bayes_loss_utility_gaussian(loss_f, mu_true, mu1, mu2, sigma1, sigma2):
x = np.linspace(-7, 7, 1000, endpoint=True)
prior = gaussian(x, mu1, sigma1)
likelihood = gaussian(x, mu2, sigma2)
mu_post, sigma_post = product_guassian(mu1, mu2, sigma1, sigma2)
posterior = gaussian(x, mu_post, sigma_post)
loss = calc_loss_func(loss_f, mu_true, x)
plot_bayes_loss_utility(x, prior, likelihood, posterior, loss, loss_f)
return None
def plot_bayes_loss_utility_uniform(loss_f, mu_true, mu, sigma):
x = np.linspace(-7, 7, 1000, endpoint=True)
prior = np.ones_like(x) / (x.max() - x.min())
likelihood = gaussian(x, mu, sigma)
posterior = likelihood
loss = calc_loss_func(loss_f, mu_true, x)
plot_bayes_loss_utility(x, prior, likelihood, posterior, loss, loss_f)
return None
def plot_bayes_loss_utility_gamma(loss_f, mu_true, alpha, beta, offset, mu, sigma):
x = np.linspace(-7, 7, 1000, endpoint=True)
prior = gamma_pdf(x-offset, alpha, beta)
likelihood = gaussian(x, mu, sigma)
posterior = np.multiply(prior, likelihood)
posterior = posterior / (posterior.sum() * (x[1] - x[0]))
loss = calc_loss_func(loss_f, mu_true, x)
plot_bayes_loss_utility(x, prior, likelihood, posterior, loss, loss_f)
return None
def plot_bayes_loss_utility_mixture(loss_f, mu_true, mu_m1, mu_m2, sigma_m1, sigma_m2, factor, mu, sigma):
x = np.linspace(-7, 7, 1000, endpoint=True)
y_1 = gaussian(x, mu_m1, sigma_m1)
y_2 = gaussian(x, mu_m2, sigma_m2)
prior = y_1 * factor + y_2 * (1.0 - factor)
likelihood = gaussian(x, mu, sigma)
posterior = np.multiply(prior, likelihood)
posterior = posterior / (posterior.sum() * (x[1] - x[0]))
loss = calc_loss_func(loss_f, mu_true, x)
plot_bayes_loss_utility(x, prior, likelihood, posterior, loss, loss_f)
return None
def plot_bayes_loss_utility(x, prior, likelihood, posterior, loss, loss_f):
mean, median, mode = calc_mean_mode_median(x, posterior)
expected_loss = calc_expected_loss(loss_f, posterior, x)
min_expected_loss = x[np.argmin(expected_loss)]
plt.figure(figsize=(12, 8))
plt.subplot(2, 2, 1)
plt.title("Prior and Likelihood")
plt.plot(x, prior, c='b', label='prior')
plt.fill_between(x, prior, color='b', alpha=0.2)
plt.plot(x, likelihood, c='r', label='likelihood')
plt.fill_between(x, likelihood, color='r', alpha=0.2)
plt.yticks([])
plt.legend(loc="upper left")
plt.xlabel('x')
plt.subplot(2, 2, 2)
plt.title("Posterior")
plt.plot(x, posterior, c='k', label='posterior')
plt.fill_between(x, posterior, color='k', alpha=0.1)
plt.axvline(mean, ls='dashed', color='red', label='Mean')
plt.axvline(median, ls='dashdot', color='blue', label='Median')
plt.axvline(mode, ls='dotted', color='green', label='Mode')
plt.legend(loc="upper left")
plt.yticks([])
plt.xlabel('x')
plt.subplot(2, 2, 3)
plt.title(loss_f)
plt.plot(x, loss, c='c', label=loss_f)
# plt.fill_between(x, loss, color='c', alpha=0.2)
plt.ylabel('loss')
plt.xlabel('x')
plt.subplot(2, 2, 4)
plt.title("expected loss")
plt.plot(x, expected_loss, c='y', label='$\mathbb{E}[L]$')
# plt.fill_between(x, expected_loss, color='y', alpha=0.2)
plt.axvline(min_expected_loss, ls='dashed', color='red', label='$Min~ \mathbb{E}[Loss]$')
plt.legend(loc="lower right")
plt.xlabel('x')
plt.ylabel('$\mathbb{E}[L]$')
plt.show()
Plotting Widgets¶
# @title Plotting Widgets
global global_loss_plot_switcher
global_loss_plot_switcher = False
def loss_plot_switcher(what_to_plot):
global global_loss_plot_switcher
if global_loss_plot_switcher:
clear_output()
else:
global_loss_plot_switcher = True
if what_to_plot == "Gaussian":
loss_f_options = Dropdown(
options=["Mean Squared Error", "Absolute Error", "Zero-One Loss"],
value="Mean Squared Error", description="Loss: ")
mu_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=-0.5, description="µ_estimate", continuous_update=True)
sigma_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_estimate", continuous_update=True)
mu_true_slider = FloatSlider(min=-3.0, max=3.0, step=0.01, value=0.0, description="µ_true", continuous_update=True)
widget_ui = HBox([VBox([loss_f_options, mu_true_slider]), VBox([mu_slider, sigma_slider])])
widget_out = interactive_output(plot_loss_utility_gaussian,
{'loss_f': loss_f_options,
'mu': mu_slider,
'sigma': sigma_slider,
'mu_true': mu_true_slider})
elif what_to_plot == "Mixture of Gaussians":
loss_f_options = Dropdown(
options=["Mean Squared Error", "Absolute Error", "Zero-One Loss"],
value="Mean Squared Error", description="Loss: ")
mu1_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=0.5, description="µ_est_p", continuous_update=True)
mu2_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=-0.5, description="µ_est_q", continuous_update=True)
sigma1_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_est_p", continuous_update=True)
sigma2_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_est_q", continuous_update=True)
factor_slider = FloatSlider(min=0.0, max=1.0, step=0.01, value=0.5, description="π", continuous_update=True)
mu_true_slider = FloatSlider(min=-3.0, max=3.0, step=0.01, value=0.0, description="µ_true", continuous_update=True)
widget_ui = HBox([VBox([loss_f_options, factor_slider, mu_true_slider]),
VBox([mu1_slider, sigma1_slider]),
VBox([mu2_slider, sigma2_slider])])
widget_out = interactive_output(plot_loss_utility_mixture,
{'mu1': mu1_slider,
'mu2': mu2_slider,
'sigma1': sigma1_slider,
'sigma2': sigma2_slider,
'factor': factor_slider,
'mu_true': mu_true_slider,
'loss_f': loss_f_options})
display(widget_ui, widget_out)
global global_plot_prior_switcher
global_plot_prior_switcher = False
def plot_prior_switcher(what_to_plot):
global global_plot_prior_switcher
if global_plot_prior_switcher:
clear_output()
else:
global_plot_prior_switcher = True
if what_to_plot == "Gaussian":
mu1_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=-0.5, description="µ_prior", continuous_update=True)
mu2_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=0.5, description="µ_likelihood", continuous_update=True)
sigma1_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_prior", continuous_update=True)
sigma2_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_likelihood", continuous_update=True)
widget_ui = HBox([VBox([mu1_slider, sigma1_slider]),
VBox([mu2_slider, sigma2_slider])])
widget_out = interactive_output(plot_utility_gaussian,
{'mu1': mu1_slider,
'mu2': mu2_slider,
'sigma1': sigma1_slider,
'sigma2': sigma2_slider,
'mu_g': fixed(1.0),
'mu_c': fixed(-1.0),
'sigma_g': fixed(0.5),
'sigma_c': fixed(value=0.5),
'plot_utility_row': fixed(False)})
elif what_to_plot == "Uniform":
mu_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=0.5, description="µ_likelihood", continuous_update=True)
sigma_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_likelihood", continuous_update=True)
widget_ui = VBox([mu_slider, sigma_slider])
widget_out = interactive_output(plot_utility_uniform,
{'mu': mu_slider,
'sigma': sigma_slider,
'mu_g': fixed(1.0),
'mu_c': fixed(-1.0),
'sigma_g': fixed(0.5),
'sigma_c': fixed(value=0.5),
'plot_utility_row': fixed(False)})
elif what_to_plot == "Gamma":
alpha_slider = FloatSlider(min=1.0, max=10.0, step=0.1, value=2.0, description="α_prior", continuous_update=True)
beta_slider = FloatSlider(min=0.5, max=2.0, step=0.01, value=1.0, description="β_prior", continuous_update=True)
# offset_slider = FloatSlider(min=-6.0, max=2.0, step=0.1, value=0.0, description="offset", continuous_update=True)
offset_slider = fixed(0.0)
mu_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=0.5, description="µ_likelihood", continuous_update=True)
sigma_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_likelihood", continuous_update=True)
gaus_label = Label(value="normal likelihood", layout=Layout(display="flex", justify_content="center"))
gamma_label = Label(value="gamma prior", layout=Layout(display="flex", justify_content="center"))
widget_ui = HBox([VBox([gamma_label, alpha_slider, beta_slider]),
VBox([gaus_label, mu_slider, sigma_slider])])
widget_out = interactive_output(plot_utility_gamma,
{'alpha': alpha_slider,
'beta': beta_slider,
'offset': offset_slider,
'mu': mu_slider,
'sigma': sigma_slider,
'mu_g': fixed(1.0),
'mu_c': fixed(-1.0),
'sigma_g': fixed(0.5),
'sigma_c': fixed(value=0.5),
'plot_utility_row': fixed(False)})
elif what_to_plot == "Mixture of Gaussians":
mu_m1_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=-0.5, description="µ_mix_p", continuous_update=True)
mu_m2_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=0.5, description="µ_mix_q", continuous_update=True)
sigma_m1_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_mix_p", continuous_update=True)
sigma_m2_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_mix_q", continuous_update=True)
factor_slider = FloatSlider(min=0.0, max=1.0, step=0.01, value=0.5, description="π", continuous_update=True)
mu_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=0.5, description="µ_likelihood", continuous_update=True)
sigma_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_likelihood", continuous_update=True)
widget_ui = HBox([VBox([mu_m1_slider, sigma_m1_slider, factor_slider]),
VBox([mu_m2_slider, sigma_m2_slider]),
VBox([mu_slider, sigma_slider])])
widget_out = interactive_output(plot_utility_mixture,
{'mu_m1': mu_m1_slider,
'mu_m2': mu_m2_slider,
'sigma_m1': sigma_m1_slider,
'sigma_m2': sigma_m2_slider,
'factor': factor_slider,
'mu': mu_slider,
'sigma': sigma_slider,
'mu_g': fixed(1.0),
'mu_c': fixed(-1.0),
'sigma_g': fixed(0.5),
'sigma_c': fixed(value=0.5),
'plot_utility_row': fixed(False)})
display(widget_ui, widget_out)
global global_plot_bayes_loss_utility_switcher
global_plot_bayes_loss_utility_switcher = False
def plot_bayes_loss_utility_switcher(what_to_plot):
global global_plot_bayes_loss_utility_switcher
if global_plot_bayes_loss_utility_switcher:
clear_output()
else:
global_plot_bayes_loss_utility_switcher = True
if what_to_plot == "Gaussian":
loss_f_options = Dropdown(
options=["Mean Squared Error", "Absolute Error", "Zero-One Loss"],
value="Mean Squared Error", description="Loss: ")
mu1_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=-0.5, description="µ_prior", continuous_update=True)
sigma1_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_prior", continuous_update=True)
mu2_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=0.5, description="µ_likelihood", continuous_update=True)
sigma2_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_likelihood", continuous_update=True)
mu_true_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=-0.5, description="µ_true", continuous_update=True)
widget_ui = HBox([VBox([loss_f_options, mu1_slider, sigma1_slider]),
VBox([mu_true_slider, mu2_slider, sigma2_slider])])
widget_out = interactive_output(plot_bayes_loss_utility_gaussian,
{'mu1': mu1_slider,
'mu2': mu2_slider,
'sigma1': sigma1_slider,
'sigma2': sigma2_slider,
'mu_true': mu_true_slider,
'loss_f': loss_f_options})
elif what_to_plot == "Uniform":
loss_f_options = Dropdown(
options=["Mean Squared Error", "Absolute Error", "Zero-One Loss"],
value="Mean Squared Error", description="Loss: ")
mu_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=0.5, description="µ_likelihood", continuous_update=True)
sigma_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_likelihood", continuous_update=True)
mu_true_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=-0.5, description="µ_true", continuous_update=True)
widget_ui = HBox([VBox([loss_f_options, mu_slider, sigma_slider]),
VBox([mu_true_slider])])
widget_out = interactive_output(plot_bayes_loss_utility_uniform,
{'mu': mu_slider,
'sigma': sigma_slider,
'mu_true': mu_true_slider,
'loss_f': loss_f_options})
elif what_to_plot == "Gamma":
loss_f_options = Dropdown(
options=["Mean Squared Error", "Absolute Error", "Zero-One Loss"],
value="Mean Squared Error", description="Loss: ")
alpha_slider = FloatSlider(min=1.0, max=10.0, step=0.1, value=2.0, description="α_prior", continuous_update=True)
beta_slider = FloatSlider(min=0.5, max=2.0, step=0.01, value=1.0, description="β_prior", continuous_update=True)
# offset_slider = FloatSlider(min=-6.0, max=2.0, step=0.1, value=0.0, description="offset", continuous_update=True)
offset_slider = fixed(0.0)
mu_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=0.5, description="µ_likelihood", continuous_update=True)
sigma_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_likelihood", continuous_update=True)
mu_true_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=-0.5, description="µ_true", continuous_update=True)
gaus_label = Label(value="normal likelihood", layout=Layout(display="flex", justify_content="center"))
gamma_label = Label(value="gamma prior", layout=Layout(display="flex", justify_content="center"))
widget_ui = HBox([VBox([loss_f_options, gamma_label, alpha_slider, beta_slider]),
VBox([mu_true_slider, gaus_label, mu_slider, sigma_slider])])
widget_out = interactive_output(plot_bayes_loss_utility_gamma,
{'alpha': alpha_slider,
'beta': beta_slider,
'offset': offset_slider,
'mu': mu_slider,
'sigma': sigma_slider,
'mu_true': mu_true_slider,
'loss_f': loss_f_options})
elif what_to_plot == "Mixture of Gaussians":
mu_m1_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=-0.5, description="µ_mix_p", continuous_update=True)
mu_m2_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=0.5, description="µ_mix_q", continuous_update=True)
sigma_m1_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_mix_p", continuous_update=True)
sigma_m2_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_mix_q", continuous_update=True)
factor_slider = FloatSlider(min=0.0, max=1.0, step=0.01, value=0.5, description="π", continuous_update=True)
mu_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=0.5, description="µ_likelihood", continuous_update=True)
sigma_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_likelihood", continuous_update=True)
mu_true_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=-0.5, description="µ_true", continuous_update=True)
loss_f_options = Dropdown(
options=["Mean Squared Error", "Absolute Error", "Zero-One Loss"],
value="Mean Squared Error", description="Loss: ")
empty_label = Label(value=" ")
widget_ui = HBox([VBox([loss_f_options, mu_m1_slider, sigma_m1_slider]),
VBox([mu_true_slider, mu_m2_slider, sigma_m2_slider]),
VBox([empty_label, mu_slider, sigma_slider])])
widget_out = interactive_output(plot_bayes_loss_utility_mixture,
{'mu_m1': mu_m1_slider,
'mu_m2': mu_m2_slider,
'sigma_m1': sigma_m1_slider,
'sigma_m2': sigma_m2_slider,
'factor': factor_slider,
'mu': mu_slider,
'sigma': sigma_slider,
'mu_true': mu_true_slider,
'loss_f': loss_f_options})
display(widget_ui, widget_out)
Helper Functions¶
# @title Helper Functions
def gaussian(x, μ, σ):
""" Compute Gaussian probability density function for given value of the
random variable, mean, and standard deviation
Args:
x (scalar): value of random variable
μ (scalar): mean of Gaussian
σ (scalar): standard deviation of Gaussian
Returns:
scalar: value of probability density function
"""
return np.exp(-((x - μ) / σ)**2 / 2) / np.sqrt(2 * np.pi * σ**2)
def gamma_pdf(x, α, β):
return gamma_distribution.pdf(x, a=α, scale=1/β)
def mvn2d(x, y, mu1, mu2, sigma1, sigma2, cov12):
mvn = multivariate_normal([mu1, mu2], [[sigma1**2, cov12], [cov12, sigma2**2]])
return mvn.pdf(np.dstack((x, y)))
def product_guassian(mu1, mu2, sigma1, sigma2):
J_1, J_2 = 1/sigma1**2, 1/sigma2**2
J_3 = J_1 + J_2
mu_prod = (J_1*mu1/J_3) + (J_2*mu2/J_3)
sigma_prod = np.sqrt(1/J_3)
return mu_prod, sigma_prod
def reverse_product(mu3, sigma3, mu1, mu2):
J_3 = 1/sigma3**2
J_1 = J_3 * (mu3 - mu2) / (mu1 - mu2)
J_2 = J_3 * (mu3 - mu1) / (mu2 - mu1)
sigma1, sigma2 = 1/np.sqrt(J_1), 1/np.sqrt(J_2)
return sigma1, sigma2
def calc_mean_mode_median(x, y):
"""
"""
pdf = y * (x[1] - x[0])
# Calc mode of an arbitrary function
mode = x[np.argmax(pdf)]
# Calc mean of an arbitrary function
mean = np.multiply(x, pdf).sum()
# Calc median of an arbitrary function
cdf = np.cumsum(pdf)
idx = np.argmin(np.abs(cdf - 0.5))
median = x[idx]
return mean, median, mode
def calc_expected_loss(loss_f, posterior, x):
dx = x[1] - x[0]
expected_loss = np.zeros_like(x)
for i in np.arange(x.shape[0]):
loss = calc_loss_func(loss_f, x[i], x) # or mse or zero_one_loss
expected_loss[i] = np.sum(loss * posterior) * dx
return expected_loss
Section 0: Introduction¶
Video 1: Introduction¶
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Introduction_Video")
Section 1: Astrocat!¶
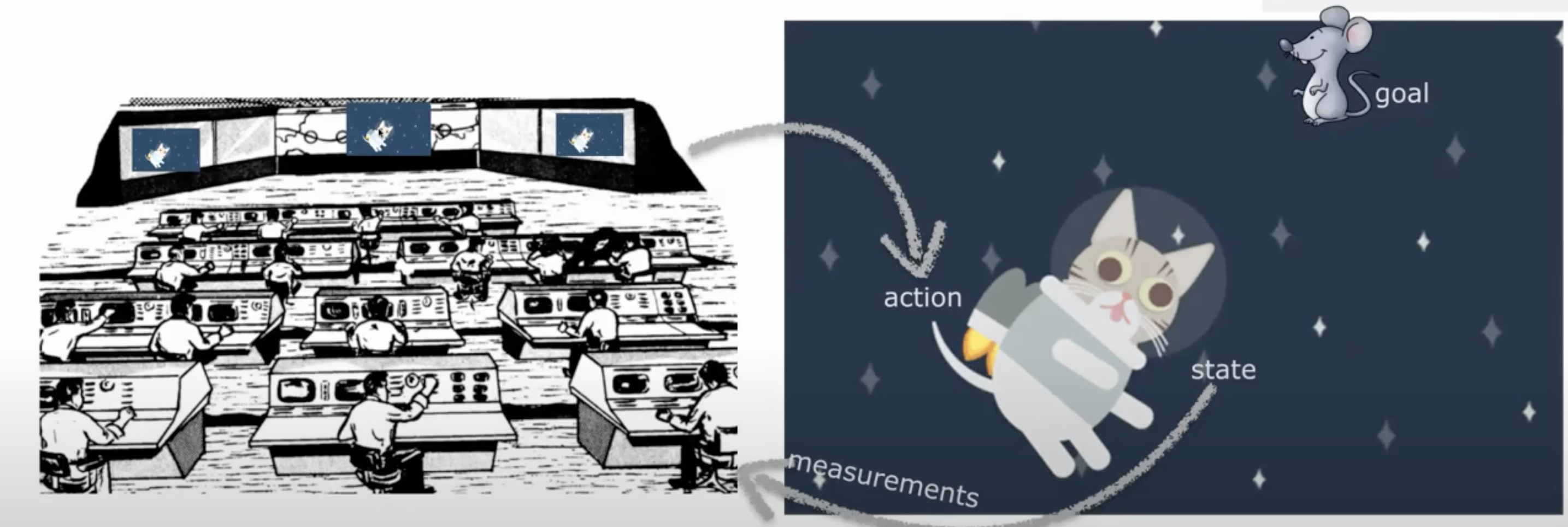
Let’s say you are a cat astronaut - Astrocat! You are navigating around space using a jetpack and your goal is to chase a mouse.
Since you are a cat, you don’t have opposable thumbs and cannot control your own jet pack. It can only be controlled by ground control back on Earth.
For them to be able to guide you, they need to know where you are. They are trying to figure out your location. They have prior knowledge of your location - they know you like to hang out near the space mouse. They can also get an unreliable quick glimpse: they are taking a measurement of the hidden state of your location.
They will try to figure out your location using Bayes rule and Bayesian decisions - as we will see throughout this tutorial.
Video 2: Astrocat¶
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Astrocat_Video")
Astrocat is in space and we are considering the position along one dimension. So, the hidden state, s, is the true location. The satellites represent potential loss, and the space mouse, gain. Using indirect measurements, you as ground control, can estimate where Astrocat is or decide where it’s likely better to send Astrocat.
Remember, in this example, you can think of yourself as a scientist trying to decide where we believe Astrocat is, how to select a point estimate (single guess of location) based on possible errors, and how to account for the uncertainty we have about the location of the satellite and the space mouse. In fact, this is the kind of problem real scientists working to control remote satellites face! However, we can also think of this as what your brain does when it wants to determine a target to make a movement or hit a tennis ball! A number of classic experiments use this kind of framing to study how optimal human decisions or movements are! Some examples are in the further reading document.
Section 2: Probability distribution of Astrocat location¶
Estimated timing to here from start of tutorial: 5 min
We are going to think first about how Ground Control should estimate his position. We won’t consider measurements yet, just how to represent the uncertainty we might have in general. We are now dealing with a continuous distribution - Astrocat’s location can be any real number. In the last tutorial, we were dealing with a discrete distribution - the fish were either on one side or the other.
So how do we represent the probability of each possible point (an infinite number) where the Astrocat could be? The Bayesian approach can be used on any probability distribution. While many variables in the world require representation using complex or unknown (e.g. empirical) distributions, we will be using the Gaussian distributions or extensions of it.
Section 2.1: The Gaussian distribution¶
Video 3: The Gaussian distribution¶
Click here for text recap of video
One distribution we will use throughout this tutorial is the Gaussian distribution, which is also sometimes called the normal distribution.
This is a special, and commonly used, distribution for a couple reasons. It is actually the focus of one of the most important theorems in statistics: the Central Limit Theorem. This theorem tells us that if you sum a large number of samples of a variable, that sum is normally distributed no matter what the original distribution over a variable was. This is a bit too in-depth for us to get into now but check out links in the Bonus for more information. Additionally, Gaussians have some really nice mathematical properties that permit simple closed-form solutions to several important problems. As we will see later in this tutorial, we can extend Gaussians to be even more flexible and well approximate other distributions using mixtures of Gaussians. In short, the Gaussian is probably the most important continous distribution to understand and use.
Gaussians have two parameters. The mean \(\mu\), which sets the location of its center. Its “scale” or spread is controlled by its standard deviation \(\sigma\) or its square, the variance \(\sigma^2\). These can be a bit easy to mix up: make sure you are careful about whether you are referring to/using standard deviation or variance.
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_The_Gaussian_distribution_Video")
The equation for a Gaussian distribution on a variable \(x\) is:
In our example, \(x\) is the location of the Astrocat in one direction. \(\mathcal{N}(\mu,\sigma^2)\) is a standard notation to refer to a Normal (Gaussian) distribution. For example, \(\mathcal{N}(0, 1)\) denotes a Gaussian distribution with mean 0 and variance 1. The exact form of this equation is not particularly intuitive, but we will see how mean and standard deviation values affect the probability distribution.
We won’t implement a Gaussian distribution in code here but please refer to the pre-reqs refresher W0D5 T1 to do this if you need further clarification.
Execute this cell to enable the function gaussian
# @markdown Execute this cell to enable the function `gaussian`
def gaussian(x, μ, σ):
return np.exp(-((x - μ) / σ)**2 / 2) / np.sqrt(2 * np.pi * σ**2)
Interactive Demo 2.1: Exploring Gaussian parameters¶
Let’s explore how the parameters of a Gaussian affect the distribution. Play with the demo below by changing the mean \(\mu\) and standard deviation \(\sigma\).
Discuss the following:
What does increasing \(\mu\) do? What does increasing \(\sigma\) do?
If you wanted to know the probability of an event happening at \(0\), can you find two different \(\mu\) and \(\sigma\) values that produce the same probability of an event at \(0\)?
How many Gaussian’s could produce the same probability at \(0\)?
Execute this cell to enable the widget
# @markdown Execute this cell to enable the widget
widget = interact(plot_gaussian,
μ = FloatSlider(min=-4.0, max=4.0, step=0.01, value=0.0, continuous_update=False),
σ = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, continuous_update=False))
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Exploring_Gaussian_parameters_Interactive_Demo_and_Discussion")
Section 2.2: Multiplying Gaussians¶
Video 4: Multiplying Gaussians¶
This video covers the multiplication of two Gaussian distributions.
Click here for text recap of video
When we multiply Gaussians, we are not multiplying random variables but the actual underlying distributions. If we multiply two Gaussian distributions, with means \(\mu_1\) and \(\mu_2\) and standard deviations \(\sigma_1\) and \(\sigma_2\), we get another Gaussian. The Gaussian resulting from the multiplication will have mean \(\mu_3\) and standard deviation \(\sigma_3\) where:
This may look confusing but keep in mind that the information in a Gaussian is the inverse of its variance: \(\frac{1}{\sigma^2}\). Basically, when multiplying Gaussians, the mean of the resulting Gaussian is a weighted average of the original means, where the weights are proportional to the amount of information of that Gaussian. The information in the resulting Gaussian is equal to the sum of informations of the original two. We’ll dive into this in the next demo.
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Multiplying_Gaussians_Video")
Interactive Demo 2.2: Multiplying Gaussians¶
We have implemented the multiplication of two Gaussians for you. Using the following widget, we are going to think about the information and combination of two Gaussians. In our case, imagine we want to find the middle location between the satellite and the space mouse. This would be the center (average) of the two locations. Because we have uncertainty, we need to weigh our uncertainty in thinking about the most likely place.
In this demo, \(\mu_{1}\) and \(\sigma_{1}\) are the mean and standard deviation of the distribution over satellite location, \(\mu_{2}\) and \(\sigma_{2}\) are the mean and standard deviation of the distribution over space mouse location, and \(\mu_{3}\) and \(\sigma_{3}\) are the mean and standard deviation of the distribution over the center location (gained by multiplying the first two).
Questions:
What is your uncertainty (how much information) do you have about \(\mu_{3}\) with \(\mu_{1} = -2, \mu_{2} = 2, \sigma_{1} = \sigma_{2} = 0.5\)?
What happens to your estimate of \(\mu_{3}\) as \(\sigma_{2} \to \infty\)? (In this case, \(\sigma\) only goes to 11… but that should be loud enough.)
What is the difference in your estimate of \(\mu_{3}\) if \(\sigma_{1} = \sigma_{2} = 11\)? What has changed from the first example?
Set \(\mu_{1} = -4, \mu_{2} = 4\) and change the \(\sigma\)s so that \(\mu_{3}\) is close to \(2\). How many \(\sigma\)s will produce the same \(\mu_{3}\)?
Continuing, if you set \(\mu_{1} = 0\), what \(\sigma\) do you need to change so \(\mu_{3} ~= 2\)?
If \(\sigma_{1} = \sigma_{2} = 0.1\), how much information do you have about the average?
Execute this cell to enable the widget
# @markdown Execute this cell to enable the widget
mu1_slider = FloatSlider(min=-5.0, max=-0.51, step=0.01, value=-2.0, description="µ_1",continuous_update=True)
mu2_slider = FloatSlider(min=0.5, max=5.01, step=0.01, value=2.0, description="µ_2",continuous_update=True)
sigma1_slider = FloatSlider(min=0.1, max=11.01, step=0.01, value=1.0, description="σ_1", continuous_update=True)
sigma2_slider = FloatSlider(min=0.1, max=11.01, step=0.01, value=1.0, description="σ_2", continuous_update=True)
distro_1_label = Label(value="Satellite", layout=Layout(display="flex", justify_content="center"))
distro_2_label = Label(value="Space Mouse", layout=Layout(display="flex", justify_content="center"))
widget_ui = HBox([VBox([distro_1_label, mu1_slider, sigma1_slider]),
VBox([distro_2_label, mu2_slider, sigma2_slider])])
widget_out = interactive_output(plot_information, {'mu1': mu1_slider,
'mu2': mu2_slider,
'sigma1': sigma1_slider,
'sigma2': sigma2_slider})
display(widget_ui, widget_out)
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Multiplying_Gaussians_Interactive_Demo_and_Discussion")
To start thinking about how we might use these concepts directly in systems neuroscience, imagine you want to know how much information is gained combining (averaging) the response of two neurons that represent locations in sensory space (think: how much information is shared by their receptive fields). You would be multiplying Gaussians!
Section 2.3: Mixtures of Gaussians¶
Video 5: Mixtures of Gaussians¶
Click here for text recap of video
What if our continuous distribution isn’t well described by a single bump? For example, what if the Astrocat is often either in one place or another - a Gaussian distribution would not capture this well! We need a multimodal distribution. Luckily, we can extend Gaussians into a mixture of Gaussians, which are more complex distributions.
In a Gaussian mixture distribution, you are essentially adding two or more weighted standard Gaussian distributions (and then normalizing so everything integrates to 1). Each standard Gaussian involved is described, as normal, by its mean and standard deviation. Additional parameters in a mixture of Gaussians are the weights you put on each Gaussian (π). The following demo should help clarify how a mixture of Gaussians relates to the standard Gaussian components. We will not cover the derivation here but you can work it out as a bonus exercise.
Mixture distributions are a common tool in Bayesian modeling and an important tool in general.
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Mixtures_of_Gaussians_Video")
Interactive Demo 2.3: Exploring Gaussian mixtures¶
We will examining a mixture of two Gaussians. We will have one weighting parameter, π, that tells us how to weigh one of the Gaussians. The other is weighted by 1 - π.
Use the following widget to experiment with the parameters of each Gaussian and the mixing weight (\(\pi\)) to understand how the mixture of Gaussian distributions behaves.
Discuss the following questions:
What does increasing the weight \(\pi\) do to the mixture distribution (dark blue)?
How can you make the two bumps of the mixture distribution further apart?
Can you make the mixture distribution have just one bump (like a Gaussian)?
Any other shapes you can make the mixture distribution resemble other than one nicely rounded bump or two separate bumps?
Execute this cell to enable the widget
# @markdown Execute this cell to enable the widget
mu1_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=-1.0, description="µ_p", continuous_update=True)
mu2_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=1.0, description="µ_q", continuous_update=True)
sigma1_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_p", continuous_update=True)
sigma2_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_q", continuous_update=True)
factor_slider = FloatSlider(min=0.1, max=0.9, step=0.01, value=0.5, description="π", continuous_update=True)
distro_1_label = Label(value="p(x)", layout=Layout(display="flex", justify_content="center"))
distro_2_label = Label(value="q(x)", layout=Layout(display="flex", justify_content="center"))
mixture_label = Label(value="mixing coefficient", layout=Layout(display="flex", justify_content="center"))
widget_ui = HBox([VBox([distro_1_label, mu1_slider, sigma1_slider]),
VBox([distro_2_label, mu2_slider, sigma2_slider]),
VBox([mixture_label, factor_slider])])
widget_out = interactive_output(gaussian_mixture, {'mu1': mu1_slider,
'mu2': mu2_slider,
'sigma1': sigma1_slider,
'sigma2': sigma2_slider,
'factor': factor_slider})
display(widget_ui, widget_out)
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Exploring_Gaussian_mixtures_Interactive_Demo_and_Discussion")
Section 3: Utility¶
Estimated timing to here from start of tutorial: 25 min
Video 6: Utility Loss Estimators¶
Click here for text recap of video
We want to know where Astrocat is. If we were asked to provide the coordinates, for example to display them for Ground Control or to note them in a log, we are not going to provide the whole probability distribution! We will give a single set of coordinates, but we first need to estimate those coordinates. Just like in the last tutorial, this may not be as easy as just what is most likely: we want to know how good or bad it is if we guess a certain location and the Astrocat is in another.
As we have seen, utility represents the gain (or if negative, loss) for us if we take a certain action for a certain value of the hidden state. In our continuous example, we need a function to be able to define the utility with respect to all possible continuous values of the state. Our action here is our guess of the Astrocat location.
We are going to explore this for the Gaussian distribution, where our estimate is \(\hat{\mu}\) and the true hidden state we are interested in is \(\mu\).
A loss function determines the “cost” (or penalty) of estimating \(\hat \mu\) when the true or correct quantity is really \(\mu\) (this is essentially the cost of the error between the true hidden state we are interested in: \(\mu\) and our estimate: \(\hat \mu\)). A loss function is equivalent to a negative utility function.
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Utility_loss_estimators_Video")
Section 3.1: Standard loss functions¶
There are lots of different possible loss functions. We will focus on three: mean-squared error where the loss is the difference between truth and estimate squared, absolute error where the loss is the absolute difference between truth and estimate, and Zero-one Loss where the loss is 1 unless we’re exactly right (the estimate equals the truth). We can represent these with the following formulas:
We will now explore how these different loss functions change our expected utility!
Check out the next cell to see the implementation of each loss in the function calc_loss_func.
Execute this cell to enable the function calc_loss_func
# @markdown Execute this cell to enable the function `calc_loss_func`
def calc_loss_func(loss_f, mu_true, x):
error = x - mu_true
if loss_f == "Mean Squared Error":
loss = (error)**2
elif loss_f == "Absolute Error":
loss = np.abs(error)
elif loss_f == "Zero-One Loss":
loss = (np.abs(error) >= 0.03).astype(np.float)
return loss
Interactive demo 3: Exploring Loss with different distributions¶
Let’s see how our loss functions interact with probability distributions to affect expected utility and consequently, the action we take.
Play with the widget below and discuss the following:
With a Gaussian distribution, does the peak of the expected utility ever change position on the x-axis for the three different losses? This peak denotes the action we would choose (the location we would guess) so in other words, would the different choices of loss function affect our action?
With a mixture of Gaussian distributions with two bumps, does the peak of the expected loss ever change position on the x-axis for the three different losses?
Find parameters for a mixture of Gaussians that results in the mean, mode, and median all being distinct (not equal to one another). With this distribution, how does the peak of the expected utility correspond to the mean/median/mode of the probability distribution for each of the three losses?
When the mixture of Gaussians has two peaks that are exactly the same height, how many modes are there?
Execute this cell to enable the widget
# @markdown Execute this cell to enable the widget
widget = interact(loss_plot_switcher,
what_to_plot = Dropdown(
options=["Gaussian", "Mixture of Gaussians"],
value="Gaussian", description="Distribution: "))
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Exploring_loss_with_different_distributions_Interactive_Demo_and_Discussion")
You can see that what coordinates you would provide for Astrocat aren’t necessarily easy to guess just from the probability distribution. You need the concept of utility/loss and a specific loss function to determine what estimate you should give.
For symmetric distributions, you will find that the mean, median and mode are the same. However, for distributions with skew, like the Gamma distribution or the Exponential distribution, these will be different. You will be able to explore more distributions as priors below.
Section 3.2: A more complex loss function¶
The loss functions we just explored were fairly simple and are often used. However, life can be complicated and in this case, Astrocat cares about both being near the space mouse and avoiding the satellites. This means we need a more complex loss function that captures this!
We know that we want to estimate Astrocat to be closer to the mouse, which is safe and desirable, but further away from the satellites, which is dangerous! So, rather than thinking about the Loss function, we will consider a generalized utility function that considers gains and losses that matter to Astrocat!
In this case, we can assume that depending on our uncertainty about Astrocat’s probable location, we may want to ‘guess’ that Astrocat is close to ‘good’ parts of space and further from ‘bad’ parts of space. We will model these utilities as Gaussian gain and loss regions–and we can assume the width of the Gaussian comes from our uncertainty over where the Space Mouse and satellite are.
Let’s explore how this works in the next interactive demo.
Interactive demo 3.2: Complicated cat costs¶
Now that we have explored Loss functions that can be used to determine both formal estimators and our expected loss given our error, we are going to see what happens to our estimates if we use a generalized utility function.
Questions:
As you change the \(\mu\) of Astrocat, what happens to the expected utility?
Can the EU be exactly zero everywhere?
Can the EU be zero in a region around Astrocat but positive and negative elsewhere?
As our uncertainty about Astrocat’s position increases, what happens to the expected utility?
Execute this cell to enable the widget
# @markdown Execute this cell to enable the widget
mu_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=0.0, description="µ", continuous_update=True)
mu_g_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=-1.0, description="µ_gain", continuous_update=True)
mu_c_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=1.0, description="µ_cost", continuous_update=True)
sigma_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ", continuous_update=True)
sigma_g_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_gain", continuous_update=True)
sigma_c_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_cost", continuous_update=True)
distro_label = Label(value="probability", layout=Layout(display="flex", justify_content="center"))
gain_label = Label(value="gain", layout=Layout(display="flex", justify_content="center"))
loss_label = Label(value="loss", layout=Layout(display="flex", justify_content="center"))
widget_ui = HBox([VBox([distro_label, mu_slider, sigma_slider]),
VBox([gain_label, mu_g_slider, sigma_g_slider]),
VBox([loss_label, mu_c_slider, sigma_c_slider])])
widget_out = interactive_output(plot_simple_utility_gaussian,
{'mu': mu_slider,
'sigma': sigma_slider,
'mu_g': mu_g_slider,
'mu_c': mu_c_slider,
'sigma_g': sigma_g_slider,
'sigma_c': sigma_c_slider})
display(widget_ui, widget_out)
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Complicated_cat_costs_Interactive_Demo_and_Discussion")
Section 4: Correlation and marginalization¶
Estimated timing to here from start of tutorial: 40 min
In this section we will explore a two dimensional Gaussian, often defined as a two-dimension vector of Gaussian random variables. This is, in essence, the joint distribution of two Gaussian random variables.
Video 7: Correlation and marginalization¶
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Correlation_and_marginalization_Video")
Section 4.1: Correlation¶
Click here for text recap of relevant part of video
If the two variables in a two dimensional Gaussian are independent, looking at one tells us nothing about the other. But what if the the two variables are correlated (covary)?
The covariance of two Gaussians with means \(\mu_X\) and \(\mu_Y\) and standard deviations \(\sigma_X\) and \(\sigma_Y\)is:
\(\mathbb{E}[\cdot]\) here denotes the expected value. So the covariance is the expected value of the random variable \(X\) minus the mean of the Gaussian distribution on \(X\) times the random variable Y minus the mean of the Gaussian distribution on \(Y\).
The correlation is the covariance normalized, so that it goes between -1 (exactly anticorrelated) to 1 (exactly correlated).
These are key concepts and while we are considering two hidden states (or two random variables), they extend to \(N\) dimensional vectors of Gaussian random variables. You will find these used all over computational neuroscience.
Interactive demo 4.1: Covarying 2D Gaussian¶
Let’s explore this 2D Gaussian (i.e. joint distribution of two Gaussians).
Use the following widget to think about the following questions:
If these variables represent hidden states we care about, what does observing one tell us about the other? How does this depend on the correlation?
How does the shape of the distribution change when we change the means? The variances? The correlation?
If we want to isolate one or the other hidden state distributions, what do we need to do? (Hint: think about Tutorial 1.)
Execute the cell to enable the widget
# @markdown Execute the cell to enable the widget
mu1_slider = FloatSlider(min=-1.0, max=1.0, step=0.01, value=0.0, description="µ_x", continuous_update=True)
# mu2_slider = FloatSlider(min=-1.0, max=1.0, step=0.01, value=0.0, description="µ_y", continuous_update=True, orientation='vertical')
mu2_slider = FloatSlider(min=-1.0, max=1.0, step=0.01, value=0.0, description="µ_y", continuous_update=True)
sigma1_slider = FloatSlider(min=0.1, max=1.5, step=0.01, value=0.5, description="σ_x", continuous_update=True)
# sigma2_slider = FloatSlider(min=0.1, max=1.5, step=0.01, value=0.5, description="σ_y", continuous_update=True, orientation='vertical')
sigma2_slider = FloatSlider(min=0.1, max=1.5, step=0.01, value=0.5, description="σ_y", continuous_update=True)
corr_slider = FloatSlider(min=-0.99, max=0.99, step=0.01, value=0.0, description="ρ", continuous_update=True)
distro1_label = Label(value="p(x)", layout=Layout(display="flex", justify_content="center"))
distro2_label = Label(value="p(y)", layout=Layout(display="flex", justify_content="center"))
corr_label = Label(value="correlation", layout=Layout(display="flex", justify_content="center"))
widget_ui = HBox([VBox([distro1_label, mu1_slider, sigma1_slider]),
VBox([corr_label, corr_slider]),
VBox([distro2_label, mu2_slider, sigma2_slider])])
widget_out = interactive_output(plot_mvn2d, {'mu1': mu1_slider,
'mu2': mu2_slider,
'sigma1': sigma1_slider,
'sigma2': sigma2_slider,
'corr': corr_slider})
display(widget_ui, widget_out)
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Covarying_2D_Gaussian_Interactive_Demo_and_Discussion")
Section 4.2: Marginalization and information¶
Click here for text recap of relevant part of video
We learned in Tutorial 1 that if we want to measure the probability of one or another variable, we need to average over the other. When we extend this to the correlated Gaussians we just played with, marginalization works the same way. Let’s say that the two variables reflect Astrocat’s position in space (in two dimensions). If we want to get our uncertainty about Astrocat’s X or Y position, we need to marginalize.
However, let’s imagine we have a measurement from one of the variables, for example X, and we want to understand the uncertainty we have in Y. We no longer want to marginalize because we know X, we don’t need to ignore it! Instead, we can calculate the conditional probability \(P(Y|X=x)\). You will explore the relationship between these two concepts in the following interactive demo.
But first, let’s remember that we can also think about the amount of uncertainty as inversely proportional to the amount of information we have about each variable. This is important, because the joint information is determined by the correlation. For our Bayesian approach, the important intuition is that we can also think about the mutual information between the prior and the likelihood following a measurement.
Interactive demo 4.2: Marginalizing 2D Gaussians¶
Use the following widget to think consider the following questions:
When is the marginal distribution the same as the conditional probability distribution? Why?
If \(\rho\) is large, how much information can we gain (in addition) looking at both variables vs just considering one?
If \(\rho\) is close to zero, but the variances of the two variables are very different, what happens to the conditional probability compared to the marginals? As \(\rho\) changes?
Execute this cell to enable the widget
# @markdown Execute this cell to enable the widget
sigma1_slider = FloatSlider(min=0.1, max=1.1, step=0.01, value=0.5, description="σ_x", continuous_update=True)
sigma2_slider = FloatSlider(min=0.1, max=1.1, step=0.01, value=0.5, description="σ_y", continuous_update=True)
c_x_slider = FloatSlider(min=-1.0, max=1.0, step=0.01, value=0.0, description="Cx", continuous_update=True)
c_y_slider = FloatSlider(min=-1.0, max=1.0, step=0.01, value=0.0, description="Cy", continuous_update=True)
corr_slider = FloatSlider(min=-1.0, max=1.0, step=0.01, value=0.0, description="ρ", continuous_update=True)
distro1_label = Label(value="x", layout=Layout(display="flex", justify_content="center"))
distro2_label = Label(value="y", layout=Layout(display="flex", justify_content="center"))
corr_label = Label(value="correlation", layout=Layout(display="flex", justify_content="center"))
widget_ui = HBox([VBox([distro1_label, sigma1_slider, c_x_slider]),
VBox([corr_label, corr_slider]),
VBox([distro2_label, sigma2_slider, c_y_slider])])
widget_out = interactive_output(plot_marginal, {'sigma1': sigma1_slider,
'sigma2': sigma2_slider,
'c_x': c_x_slider,
'c_y': c_y_slider,
'corr': corr_slider})
display(widget_ui, widget_out)
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Marginalization_and_information_Interactive_Demo_and_Discussion")
Section 5: Bayes’ theorem for continuous distributions¶
Estimated timing to here from start of tutorial: 55 min
Video 8: Posterior Beliefs¶
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Posterior_beliefs_Video")
The continuous case allows us to consider how the shape of the posterior distribution can differ from the prior. The Gaussian case is the most fundamental, but asymmetric priors (or likelihoods) and posteriors allow us to see how the mean, median and mode can be affected differently when we apply Bayes’ theorem.
Section 5.1: The Gaussian example¶
Bayes’ rule tells us how to combine two sources of information: the prior (e.g., a noisy representation of Ground Control’s expectations about where Astrocat is) and the likelihood (e.g., a noisy representation of the Astrocat after taking a measurement), to obtain a posterior distribution (our belief distribution) taking into account both pieces of information. Remember Bayes’ rule:
We will look at what happens when both the prior and likelihood are Gaussians. In these equations, \(\mathcal{N}(\mu,\sigma^2)\) denotes a Gaussian distribution with parameters \(\mu\) and \(\sigma^2\):
When both the prior and likelihood are Gaussians, Bayes Rule translates into the following form:
We get the parameters of the posterior from multiplying the Gaussians, just as we did in Secton 2.2.
Interactive Demo 5.1: Gaussian Bayes¶
Let’s consider the following questions using the following interactive demo:
For a Gaussian posterior, explain how the information seems to be combining. (Hint: think about the prior exercises!)
What is the difference between the posterior here and the Gaussian that represented the average of two Gaussians in the exercise above?
How should we think about the relative weighting of information between the prior and posterior?
Execute this cell to enable the widget
# @markdown Execute this cell to enable the widget
mu1_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=-0.5, description="µ_prior", continuous_update=True)
mu2_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=0.5, description="µ_likelihood", continuous_update=True)
sigma1_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_prior", continuous_update=True)
sigma2_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_likelihood", continuous_update=True)
distro1_label = Label(value="prior distribution", layout=Layout(display="flex", justify_content="center"))
distro2_label = Label(value="likelihood distribution", layout=Layout(display="flex", justify_content="center"))
widget_ui = HBox([VBox([distro1_label, mu1_slider, sigma1_slider]),
VBox([distro2_label, mu2_slider, sigma2_slider])])
widget_out = interactive_output(plot_bayes, {'mu1': mu1_slider,
'mu2': mu2_slider,
'sigma1': sigma1_slider,
'sigma2': sigma2_slider})
display(widget_ui, widget_out)
Section 5.2: Exploring priors¶
Interactive Demo 5.2: Prior exploration¶
What would happen if we had a different prior distribution for Astrocat’s location? Bayes’ Rule works exactly the same way if our prior is not a Guassian (though the analytical solution may be far more complex or impossible). Let’s look at how the posterior behaves if we have a different prior over Astrocat’s location.
Consider the following questions:
Why does the posterior not look Gaussian when you use a non-Gaussian prior?
What does having a flat prior mean?
How does the Gamma prior behave differently than the others?
From what you know, can you imagine the likelihood being something other than a Gaussian?
Execute this cell to enable the widget
# @markdown Execute this cell to enable the widget
widget = interact(plot_prior_switcher,
what_to_plot = Dropdown(
options=["Gaussian", "Mixture of Gaussians", "Uniform", "Gamma"],
value="Gaussian", description="Prior: "))
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Prior_exploration_Interactive_Demo_and_Discussion")
Section 6: Bayesian decisions¶
Estimated timing to here from start of tutorial: 1 hour, 15 min
Video 9: Bayesian Decisions¶
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Bayesian_Decisions_Video")
Bayesian decisions in continuous dimensions are the same as we saw in Tutorial 1 for the binary case. The only difference is that now, our Expected Utility is calculated using an integral and all of our probability distributions are continuous.
Section 6.1: Bayesian estimation on the posterior¶
Now that we understand that the posterior can be something other than a Gaussian, let’s revisit Loss functions. In this case, we can see that the posterior can take many forms.
Interactive Demo 6.1: Standard loss functions with various priors¶
Questions:
If we have a bi-modal prior, how do the different loss functions potentially inform us differently about what we learn?
Why do the different loss functions behave differently with respect to the shape of the posterior? When do they produce different expected loss?
For the mixture of Gaussians, describe the situations where the expected loss will look different from the Gaussian case.
Execute this cell to enable the widget
# @markdown Execute this cell to enable the widget
widget = interact(plot_bayes_loss_utility_switcher,
what_to_plot = Dropdown(
options=["Gaussian", "Mixture of Gaussians", "Uniform", "Gamma"],
value="Gaussian", description="Prior: "))
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Standard_loss_functions_with_various_priors_Interactive_Demo_and_Discussion")
Section 6.2: Bayesian decisions¶
Finally, we can combine everything we have learned so far!
Now, let’s imagine we have just received a new measurement of Astrocat’s location. We need to think about how we want to decide where Astrocat is, so that we can decide how far to tell Astrocat to move. However, we want to account for the satellite and Space Mouse location in this estimation. If we make an error towards the satellite, it’s worse than towards Space Mouse. So, we will use our more complex utility function from Section 3.2.
Interactive Demo 6.2: Complicated cat costs with various priors¶
Questions:
If you have a weak prior and likelihood, how much are you relying on the utility function to guide your estimation?
If you get a good measurement, that is a likelihood with low variance, how much does this help?
Which of the factors are most important in making your decision?
Execute this cell to enable the widget
# @markdown Execute this cell to enable the widget
mu1_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=-0.5, description="µ_prior", continuous_update=True)
mu2_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=0.5, description="µ_likelihood", continuous_update=True)
sigma1_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_prior", continuous_update=True)
sigma2_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_likelihood", continuous_update=True)
mu_g_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=0.0, description="µ_gain", continuous_update=True)
sigma_g_slider = FloatSlider(min=0.1, max=2.0, step=0.01, value=0.5, description="σ_gain", continuous_update=True)
dist_label = Label(value="loss distance: µ1_c - µ2_c", layout=Layout(display="flex", justify_content="center"))
loc_label = Label(value="loss center: (µ1_c + µ2_c) / 2", layout=Layout(display="flex", justify_content="center"))
mu_dist_slider = FloatSlider(min=0.0, max=8.0, step=0.01, value=4.0, description="distance", continuous_update=True)
mu_loc_slider = FloatSlider(min=-4.0, max=4.0, step=0.01, value=0.0, description="center", continuous_update=True)
widget_ui = HBox([VBox([mu1_slider, sigma1_slider, mu2_slider, sigma2_slider]),
VBox([dist_label, mu_dist_slider, loc_label, mu_loc_slider]),
VBox([mu_g_slider, sigma_g_slider])])
widget_out = interactive_output(plot_utility_mixture_dist,
{'mu1': mu1_slider,
'sigma1': sigma1_slider,
'mu2': mu2_slider,
'sigma2': sigma2_slider,
'mu_g': mu_g_slider,
'sigma_g': sigma_g_slider,
'mu_dist': mu_dist_slider,
'mu_loc': mu_loc_slider,
'plot_utility_row': fixed(True)})
display(widget_ui, widget_out)
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Complicated_cat_costs_with_various_priors_Interactive_Demo_and_Discussion")
Summary¶
Estimated timing of tutorial: 1 hour, 35 minutes
In this tutorial, you extended your exploration of Bayes Rule and the Bayesian approach in the context of finding and choosing a location for Astrocat.
Specifically, we covered:
The Gaussian distribution and its properties
That the likelihood is the probability of the measurement given some hidden state
Information shared between Gaussians (via multiplication of PDFs and via two-dimensional distributions)
That how the prior and likelihood interact to create the posterior, the probability of the hidden state given a measurement, depends on how they covary
That utility is the gain from each action and state pair, and the expected utility for an action is the sum of the utility for all state pairs, weighted by the probability of that state happening. You can then choose the action with highest expected utility.

