Tutorial 2: Principal Component Analysis
Contents
Tutorial 2: Principal Component Analysis¶
Week 1, Day 4: Dimensionality Reduction
By Neuromatch Academy
Content creators: Alex Cayco Gajic, John Murray
Content reviewers: Roozbeh Farhoudi, Matt Krause, Spiros Chavlis, Richard Gao, Michael Waskom, Siddharth Suresh, Natalie Schaworonkow, Ella Batty
Production editor: Spiros Chavlis
Tutorial Objectives¶
Estimated timing of tutorial: 45 minutes
In this notebook we’ll learn how to perform PCA by projecting the data onto the eigenvectors of its covariance matrix.
Overview:
Calculate the eigenvectors of the sample covariance matrix.
Perform PCA by projecting data onto the eigenvectors of the covariance matrix.
Plot and explore the eigenvalues.
To quickly refresh your knowledge of eigenvalues and eigenvectors, you can watch this short video (4 minutes) for a geometrical explanation. For a deeper understanding, this in-depth video (17 minutes) provides an excellent basis and is beautifully illustrated.
Setup¶
⚠ Experimental LLM-enhanced tutorial ⚠
This notebook includes Neuromatch’s experimental Chatify 🤖 functionality. The Chatify notebook extension adds support for a large language model-based “coding tutor” to the materials. The tutor provides automatically generated text to help explain any code cell in this notebook.
Note that using Chatify may cause breaking changes and/or provide incorrect or misleading information. If you wish to proceed by installing and enabling the Chatify extension, you should run the next two code blocks (hidden by default). If you do not want to use this experimental version of the Neuromatch materials, please use the stable materials instead.
To use the Chatify helper, insert the %%explain magic command at the start of any code cell and then run it (shift + enter) to access an interface for receiving LLM-based assitance. You can then select different options from the dropdown menus depending on what sort of assitance you want. To disable Chatify and run the code block as usual, simply delete the %%explain command and re-run the cell.
Note that, by default, all of Chatify’s responses are generated locally. This often takes several minutes per response. Once you click the “Submit request” button, just be patient– stuff is happening even if you can’t see it right away!
Thanks for giving Chatify a try! Love it? Hate it? Either way, we’d love to hear from you about your Chatify experience! Please consider filling out our brief survey to provide feedback and help us make Chatify more awesome!
Run the next two cells to install and configure Chatify…
%pip install -q davos
import davos
davos.config.suppress_stdout = True
Note: you may need to restart the kernel to use updated packages.
smuggle chatify # pip: git+https://github.com/ContextLab/chatify.git
%load_ext chatify
Downloading and initializing model; this may take a few minutes...
llama.cpp: loading model from /home/runner/.cache/huggingface/hub/models--TheBloke--Llama-2-7B-Chat-GGML/snapshots/501a3c8182cd256a859888fff4e838c049d5d7f6/llama-2-7b-chat.ggmlv3.q5_1.bin
llama_model_load_internal: format = ggjt v3 (latest)
llama_model_load_internal: n_vocab = 32000
llama_model_load_internal: n_ctx = 512
llama_model_load_internal: n_embd = 4096
llama_model_load_internal: n_mult = 256
llama_model_load_internal: n_head = 32
llama_model_load_internal: n_layer = 32
llama_model_load_internal: n_rot = 128
llama_model_load_internal: freq_base = 10000.0
llama_model_load_internal: freq_scale = 1
llama_model_load_internal: ftype = 9 (mostly Q5_1)
llama_model_load_internal: n_ff = 11008
llama_model_load_internal: model size = 7B
llama_model_load_internal: ggml ctx size = 0.08 MB
llama_model_load_internal: mem required = 6390.60 MB (+ 1026.00 MB per state)
llama_new_context_with_model: kv self size = 256.00 MB
AVX = 1 | AVX2 = 1 | AVX512 = 1 | AVX512_VBMI = 0 | AVX512_VNNI = 0 | FMA = 1 | NEON = 0 | ARM_FMA = 0 | F16C = 1 | FP16_VA = 0 | WASM_SIMD = 0 | BLAS = 0 | SSE3 = 1 | VSX = 0 |
Install and import feedback gadget¶
# @title Install and import feedback gadget
!pip3 install vibecheck datatops --quiet
from vibecheck import DatatopsContentReviewContainer
def content_review(notebook_section: str):
return DatatopsContentReviewContainer(
"", # No text prompt
notebook_section,
{
"url": "https://pmyvdlilci.execute-api.us-east-1.amazonaws.com/klab",
"name": "neuromatch_cn",
"user_key": "y1x3mpx5",
},
).render()
feedback_prefix = "W1D4_T2"
# Imports
import numpy as np
import matplotlib.pyplot as plt
Figure Settings¶
# @title Figure Settings
import logging
logging.getLogger('matplotlib.font_manager').disabled = True
import ipywidgets as widgets # interactive display
%config InlineBackend.figure_format = 'retina'
plt.style.use("https://raw.githubusercontent.com/NeuromatchAcademy/course-content/main/nma.mplstyle")
Plotting Functions¶
# @title Plotting Functions
def plot_eigenvalues(evals):
"""
Plots eigenvalues.
Args:
(numpy array of floats) : Vector of eigenvalues
Returns:
Nothing.
"""
plt.figure(figsize=(4, 4))
plt.plot(np.arange(1, len(evals) + 1), evals, 'o-k')
plt.xlabel('Component')
plt.ylabel('Eigenvalue')
plt.title('Scree plot')
plt.xticks(np.arange(1, len(evals) + 1))
plt.ylim([0, 2.5])
def plot_data(X):
"""
Plots bivariate data. Includes a plot of each random variable, and a scatter
scatter plot of their joint activity. The title indicates the sample
correlation calculated from the data.
Args:
X (numpy array of floats) : Data matrix each column corresponds to a
different random variable
Returns:
Nothing.
"""
fig = plt.figure(figsize=[8, 4])
gs = fig.add_gridspec(2, 2)
ax1 = fig.add_subplot(gs[0, 0])
ax1.plot(X[:, 0], color='k')
plt.ylabel('Neuron 1')
ax2 = fig.add_subplot(gs[1, 0])
ax2.plot(X[:, 1], color='k')
plt.xlabel('Sample Number (sorted)')
plt.ylabel('Neuron 2')
ax3 = fig.add_subplot(gs[:, 1])
ax3.plot(X[:, 0], X[:, 1], '.', markerfacecolor=[.5, .5, .5],
markeredgewidth=0)
ax3.axis('equal')
plt.xlabel('Neuron 1 activity')
plt.ylabel('Neuron 2 activity')
plt.title('Sample corr: {:.1f}'.format(np.corrcoef(X[:, 0], X[:, 1])[0, 1]))
plt.show()
def plot_data_new_basis(Y):
"""
Plots bivariate data after transformation to new bases. Similar to plot_data
but with colors corresponding to projections onto basis 1 (red) and
basis 2 (blue).
The title indicates the sample correlation calculated from the data.
Note that samples are re-sorted in ascending order for the first random
variable.
Args:
Y (numpy array of floats) : Data matrix in new basis each column
corresponds to a different random variable
Returns:
Nothing.
"""
fig = plt.figure(figsize=[8, 4])
gs = fig.add_gridspec(2, 2)
ax1 = fig.add_subplot(gs[0, 0])
ax1.plot(Y[:, 0], 'r')
plt.ylabel('Projection \n basis vector 1')
ax2 = fig.add_subplot(gs[1, 0])
ax2.plot(Y[:, 1], 'b')
plt.xlabel('Sample number')
plt.ylabel('Projection \n basis vector 2')
ax3 = fig.add_subplot(gs[:, 1])
ax3.plot(Y[:, 0], Y[:, 1], '.', color=[.5, .5, .5])
ax3.axis('equal')
plt.xlabel('Projection basis vector 1')
plt.ylabel('Projection basis vector 2')
plt.title('Sample corr: {:.1f}'.format(np.corrcoef(Y[:, 0], Y[:, 1])[0, 1]))
plt.show()
def plot_basis_vectors(X, W):
"""
Plots bivariate data as well as new basis vectors.
Args:
X (numpy array of floats) : Data matrix each column corresponds to a
different random variable
W (numpy array of floats) : Square matrix representing new orthonormal
basis each column represents a basis vector
Returns:
Nothing.
"""
plt.figure(figsize=[4, 4])
plt.plot(X[:, 0], X[:, 1], '.', color=[.5, .5, .5], label='Data')
plt.axis('equal')
plt.xlabel('Neuron 1 activity')
plt.ylabel('Neuron 2 activity')
plt.plot([0, W[0, 0]], [0, W[1, 0]], color='r', linewidth=3,
label='Basis vector 1')
plt.plot([0, W[0, 1]], [0, W[1, 1]], color='b', linewidth=3,
label='Basis vector 2')
plt.legend()
plt.show()
Helper functions¶
# @title Helper functions
def sort_evals_descending(evals, evectors):
"""
Sorts eigenvalues and eigenvectors in decreasing order. Also aligns first two
eigenvectors to be in first two quadrants (if 2D).
Args:
evals (numpy array of floats) : Vector of eigenvalues
evectors (numpy array of floats) : Corresponding matrix of eigenvectors
each column corresponds to a different
eigenvalue
Returns:
(numpy array of floats) : Vector of eigenvalues after sorting
(numpy array of floats) : Matrix of eigenvectors after sorting
"""
index = np.flip(np.argsort(evals))
evals = evals[index]
evectors = evectors[:, index]
if evals.shape[0] == 2:
if np.arccos(np.matmul(evectors[:, 0],
1 / np.sqrt(2) * np.array([1, 1]))) > np.pi / 2:
evectors[:, 0] = -evectors[:, 0]
if np.arccos(np.matmul(evectors[:, 1],
1 / np.sqrt(2) * np.array([-1, 1]))) > np.pi / 2:
evectors[:, 1] = -evectors[:, 1]
return evals, evectors
def get_data(cov_matrix):
"""
Returns a matrix of 1000 samples from a bivariate, zero-mean Gaussian
Note that samples are sorted in ascending order for the first random
variable.
Args:
var_1 (scalar) : variance of the first random variable
var_2 (scalar) : variance of the second random variable
cov_matrix (numpy array of floats) : desired covariance matrix
Returns:
(numpy array of floats) : samples from the bivariate Gaussian,
with each column corresponding to a
different random variable
"""
mean = np.array([0, 0])
X = np.random.multivariate_normal(mean, cov_matrix, size=1000)
indices_for_sorting = np.argsort(X[:, 0])
X = X[indices_for_sorting, :]
return X
def calculate_cov_matrix(var_1, var_2, corr_coef):
"""
Calculates the covariance matrix based on the variances and
correlation coefficient.
Args:
var_1 (scalar) : variance of the first random variable
var_2 (scalar) : variance of the second random variable
corr_coef (scalar) : correlation coefficient
Returns:
(numpy array of floats) : covariance matrix
"""
cov = corr_coef * np.sqrt(var_1 * var_2)
cov_matrix = np.array([[var_1, cov], [cov, var_2]])
return cov_matrix
def define_orthonormal_basis(u):
"""
Calculates an orthonormal basis given an arbitrary vector u.
Args:
u (numpy array of floats) : arbitrary 2D vector used for new basis
Returns:
(numpy array of floats) : new orthonormal basis columns correspond to
basis vectors
"""
u = u / np.sqrt(u[0] ** 2 + u[1] ** 2)
w = np.array([-u[1], u[0]])
W = np.column_stack((u, w))
return W
def change_of_basis(X, W):
"""
Projects data onto a new basis.
Args:
X (numpy array of floats) : Data matrix each column corresponding to a
different random variable
W (numpy array of floats) : new orthonormal basis columns correspond to
basis vectors
Returns:
(numpy array of floats) : Data matrix expressed in new basis
"""
Y = np.matmul(X, W)
return Y
Video 1: PCA¶
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_PCA_Video")
Section 1: Calculate the eigenvectors of the the sample covariance matrix¶
As we saw in the lecture, PCA represents data in a new orthonormal basis defined by the eigenvectors of the covariance matrix. Remember that in the previous tutorial, we generated bivariate normal data with a specified covariance matrix \(\bf \Sigma\), whose \((i,j)\)th element is:
However, in real life we don’t have access to this ground-truth covariance matrix. To get around this, we can use the sample covariance matrix, \(\bf\hat\Sigma\), which is calculated directly from the data. The \((i,j)\)th element of the sample covariance matrix is:
where \({\bf x}_i = [ x_i(1), x_i(2), \dots,x_i(N_\text{samples})]^\top\) is a column vector representing all measurements of neuron \(i\), and \(\bar {\bf x}_i\) is the mean of neuron \(i\) across samples:
If we assume that the data has already been mean-subtracted, then we can write the sample covariance matrix in a much simpler matrix form:
where \(\bf X\) is the full data matrix (each column of \(\bf X\) is a different \(\bf x_i\)).
Coding Exercise 1.1: Calculate the covariance matrix¶
Before calculating the eigenvectors, you must first calculate the sample covariance matrix.
Steps
Complete the function
get_sample_cov_matrixby first subtracting the sample mean of the data, then calculate \(\bf \hat \Sigma\) using the equation above.Use
get_datato generate bivariate normal data, and calculate the sample covariance matrix with your finishedget_sample_cov_matrix. Compare this estimate to the true covariate matrix usingcalculate_cov_matrix. You’ve seen bothget_dataandcalculate_cov_matrixin Tutorial 1.
help(get_data)
help(calculate_cov_matrix)
Help on function get_data in module __main__:
get_data(cov_matrix)
Returns a matrix of 1000 samples from a bivariate, zero-mean Gaussian
Note that samples are sorted in ascending order for the first random
variable.
Args:
var_1 (scalar) : variance of the first random variable
var_2 (scalar) : variance of the second random variable
cov_matrix (numpy array of floats) : desired covariance matrix
Returns:
(numpy array of floats) : samples from the bivariate Gaussian,
with each column corresponding to a
different random variable
Help on function calculate_cov_matrix in module __main__:
calculate_cov_matrix(var_1, var_2, corr_coef)
Calculates the covariance matrix based on the variances and
correlation coefficient.
Args:
var_1 (scalar) : variance of the first random variable
var_2 (scalar) : variance of the second random variable
corr_coef (scalar) : correlation coefficient
Returns:
(numpy array of floats) : covariance matrix
def get_sample_cov_matrix(X):
"""
Returns the sample covariance matrix of data X
Args:
X (numpy array of floats) : Data matrix each column corresponds to a
different random variable
Returns:
(numpy array of floats) : Covariance matrix
"""
#################################################
## TODO for students: calculate the covariance matrix
# Fill out function and remove
raise NotImplementedError("Student exercise: calculate the covariance matrix!")
#################################################
# Subtract the mean of X
X = ...
# Calculate the covariance matrix (hint: use np.matmul)
cov_matrix = ...
return cov_matrix
# Set parameters
np.random.seed(2020) # set random seed
variance_1 = 1
variance_2 = 1
corr_coef = 0.8
# Calculate covariance matrix
cov_matrix = calculate_cov_matrix(variance_1, variance_2, corr_coef)
print(cov_matrix)
# Generate data with that covariance matrix
X = get_data(cov_matrix)
# Get sample covariance matrix
sample_cov_matrix = get_sample_cov_matrix(X)
print(sample_cov_matrix)
SAMPLE OUTPUT
[[1. 0.8]
[0.8 1. ]]
[[0.99315313 0.82347589]
[0.82347589 1.01281397]]
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Calculate_the_covariance_matrix_Exercise")
We get a sample covariance matrix that doesn’t look too far off from the true covariance matrix, so that’s a good sign!
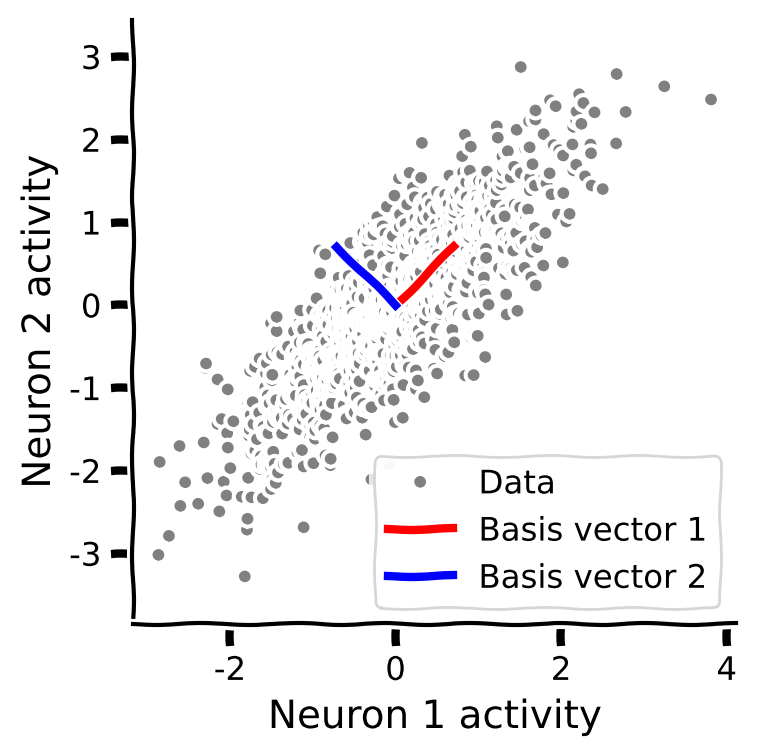
Coding Exercise 1.2: Eigenvectors of the covariance matrix¶
Next you will calculate the eigenvectors of the covariance matrix. Plot them along with the data to check that they align with the geometry of the data.
Steps:
Calculate the eigenvalues and eigenvectors of the sample covariance matrix. (Hint: use
np.linalg.eigh, which finds the eigenvalues of a symmetric matrix).Use the provided code to sort the eigenvalues in descending order.
Plot the eigenvectors on a scatter plot of the data, using the function
plot_basis_vectors.
help(sort_evals_descending)
Help on function sort_evals_descending in module __main__:
sort_evals_descending(evals, evectors)
Sorts eigenvalues and eigenvectors in decreasing order. Also aligns first two
eigenvectors to be in first two quadrants (if 2D).
Args:
evals (numpy array of floats) : Vector of eigenvalues
evectors (numpy array of floats) : Corresponding matrix of eigenvectors
each column corresponds to a different
eigenvalue
Returns:
(numpy array of floats) : Vector of eigenvalues after sorting
(numpy array of floats) : Matrix of eigenvectors after sorting
#################################################
## TODO for students
# Fill out function and remove
raise NotImplementedError("Student exercise: calculate and sort eigenvalues")
#################################################
# Calculate the eigenvalues and eigenvectors
evals, evectors = ...
# Sort the eigenvalues in descending order
evals, evectors = ...
# Visualize
plot_basis_vectors(X, evectors)
Example output:
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Eigenvectors_of_the_covariance_matrix_Exercise")
Section 2: Perform PCA by projecting data onto the eigenvectors¶
Estimated timing to here from start of tutorial: 25 min
To perform PCA, we will project the data onto the eigenvectors of the covariance matrix, i.e.,:
where \(\bf S\) is an \(N_\text{samples} \times N\) matrix representing the projected data (also called scores), and \(\bf W\) is an \(N\times N\) orthogonal matrix, each of whose columns represents the eigenvectors of the covariance matrix (also called weights or loadings).
Coding Exercise 2: PCA implementation¶
You will now perform PCA on the data using the intuition and functions you have developed so far. Fill in the function below to carry out the steps to perform PCA by projecting the data onto the eigenvectors of its covariance matrix.
Steps:
First subtract the mean and calculate the sample covariance matrix.
Then find the eigenvalues and eigenvectors and sort them in descending order.
Finally project the mean-centered data onto the eigenvectors.
help(change_of_basis)
Help on function change_of_basis in module __main__:
change_of_basis(X, W)
Projects data onto a new basis.
Args:
X (numpy array of floats) : Data matrix each column corresponding to a
different random variable
W (numpy array of floats) : new orthonormal basis columns correspond to
basis vectors
Returns:
(numpy array of floats) : Data matrix expressed in new basis
def pca(X):
"""
Sorts eigenvalues and eigenvectors in decreasing order.
Args:
X (numpy array of floats): Data matrix each column corresponds to a
different random variable
Returns:
(numpy array of floats) : Data projected onto the new basis
(numpy array of floats) : Vector of eigenvalues
(numpy array of floats) : Corresponding matrix of eigenvectors
"""
#################################################
## TODO for students: calculate the covariance matrix
# Fill out function and remove
raise NotImplementedError("Student exercise: sort eigenvalues/eigenvectors!")
#################################################
# Calculate the sample covariance matrix
cov_matrix = ...
# Calculate the eigenvalues and eigenvectors
evals, evectors = ...
# Sort the eigenvalues in descending order
evals, evectors = ...
# Project the data onto the new eigenvector basis
score = ...
return score, evectors, evals
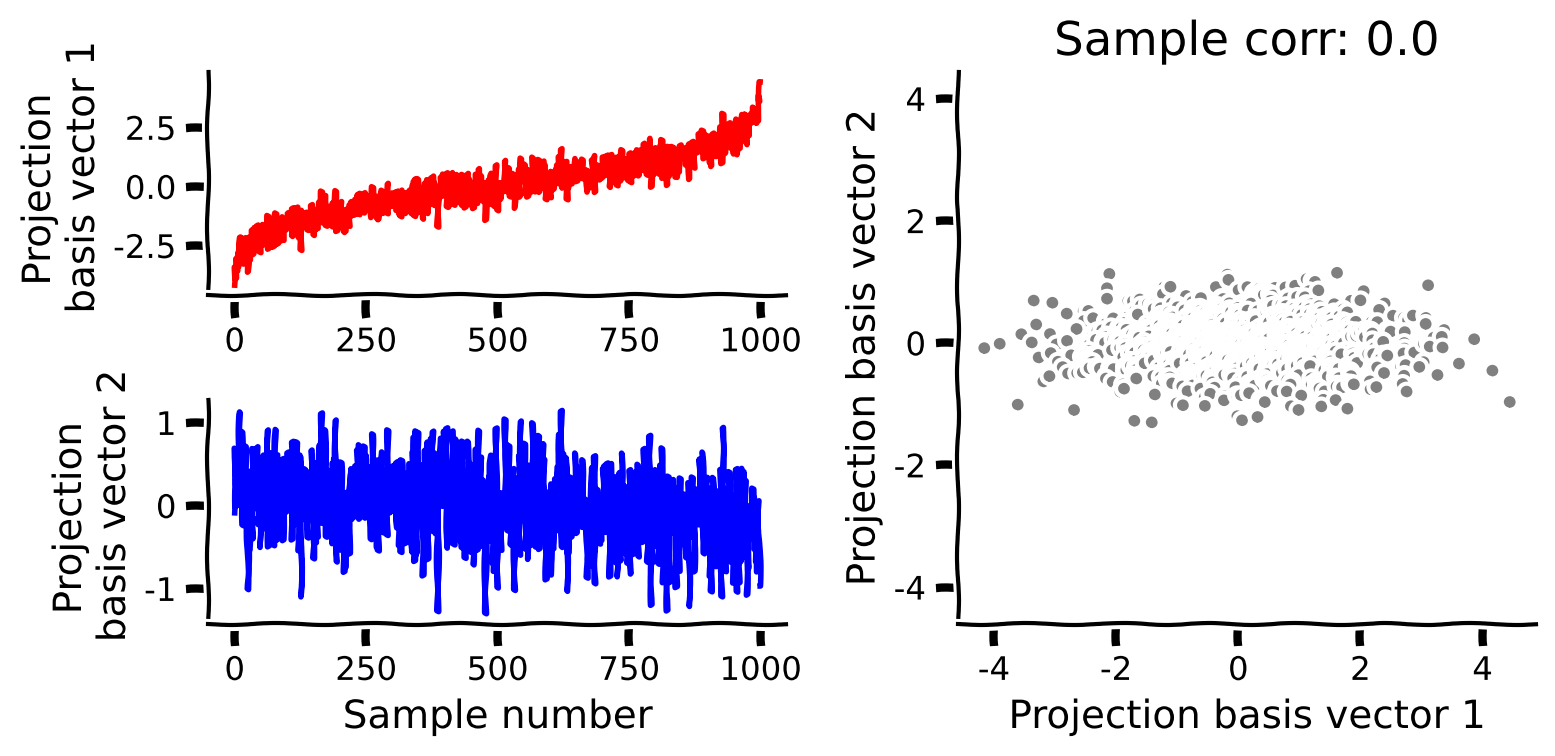
# Perform PCA on the data matrix X
score, evectors, evals = pca(X)
# Plot the data projected into the new basis
plot_data_new_basis(score)
Example output:
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_PCA_implementation_Exercise")
Finally, we will examine the eigenvalues of the covariance matrix. Remember that each eigenvalue describes the variance of the data projected onto its corresponding eigenvector. This is an important concept because it allows us to rank the PCA basis vectors based on how much variance each one can capture. First run the code below to plot the eigenvalues (sometimes called the “scree plot”). Which eigenvalue is larger?
plot_eigenvalues(evals)
Interactive Demo 2: Exploration of the correlation coefficient¶
Run the following cell and use the slider to change the correlation coefficient in the data. You should see the scree plot and the plot of basis vectors updated.
What happens to the eigenvalues as you change the correlation coefficient?
Can you find a value for which both eigenvalues are equal?
Can you find a value for which only one eigenvalue is nonzero?
Make sure you execute this cell to enable the widget!
# @markdown Make sure you execute this cell to enable the widget!
def refresh(corr_coef=.8):
cov_matrix = calculate_cov_matrix(variance_1, variance_2, corr_coef)
X = get_data(cov_matrix)
score, evectors, evals = pca(X)
plot_eigenvalues(evals)
plot_basis_vectors(X, evectors)
_ = widgets.interact(refresh, corr_coef=(-1, 1, .1))
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Exploration_of_the_correlation_coefficient_Interactive_Demo_and_Discussion")
Summary¶
Estimated timing of tutorial: 45 minutes
In this tutorial, we learned that the goal of PCA is to find an orthonormal basis capturing the directions of maximum variance of the data. More precisely, the \(i\)th basis vector is the direction that maximizes the projected variance, while being orthogonal to all previous basis vectors. Mathematically, these basis vectors are the eigenvectors of the covariance matrix (also called loadings).
PCA also has the useful property that the projected data (scores) are uncorrelated.
The projected variance along each basis vector is given by its corresponding eigenvalue. This is important because it allows us to rank the “importance” of each basis vector in terms of how much of the data variability it explains. An eigenvalue of zero means there is no variation along that direction so it can be dropped without losing any information about the original data.
In the next tutorial, we will use this property to reduce the dimensionality of high-dimensional data.

