Tutorial 2: Time Series, Global Averages, and Scenario Comparison
Contents
Tutorial 2: Time Series, Global Averages, and Scenario Comparison#
Week 2, Day 1, Future Climate: The Physical Basis
Content creators: Brodie Pearson, Julius Busecke, Tom Nicholas
Content reviewers: Younkap Nina Duplex, Zahra Khodakaramimaghsoud, Sloane Garelick, Peter Ohue, Jenna Pearson, Derick Temfack, Peizhen Yang, Cheng Zhang, Chi Zhang, Ohad Zivan
Content editors: Jenna Pearson, Ohad Zivan, Chi Zhang
Production editors: Wesley Banfield, Jenna Pearson, Chi Zhang, Ohad Zivan
Our 2023 Sponsors: NASA TOPS, Google DeepMind, and CMIP
Tutorial Objectives#
In this tutorial, we will expand to look at data from three CMIP6/ScenarioMIP experiments (historical, SSP1-2.6 and SSP5-8.5). Our aim will be to calculate the global mean SST for these 3 experiments, taking into account the spatially-varying size of the model’s grid cells (i.e., calculating a weighted mean).
By the end of this tutorial, you’ll be able to:
Load and analyze CMIP6 SST data from different experiments.
Understand the difference between historical and future emission scenarios.
Calculate the global mean SST from gridded model data.
Apply the concept of weighted mean to account for varying grid cell sizes in Earth System Models.
Setup#
# installations ( uncomment and run this cell ONLY when using google colab or kaggle )
# !pip install condacolab &> /dev/null
# import condacolab
# condacolab.install()
# # Install all packages in one call (+ use mamba instead of conda), this must in one line or code will fail
# !mamba install xarray-datatree intake-esm gcsfs xmip aiohttp nc-time-axis cf_xarray xmip xarrayutils &> /dev/null
# imports
import time
tic = time.time()
import intake
import numpy as np
import matplotlib.pyplot as plt
import xarray as xr
from xmip.preprocessing import combined_preprocessing
from xarrayutils.plotting import shaded_line_plot
from datatree import DataTree
from xmip.postprocessing import _parse_metric
# @title Figure settings
import ipywidgets as widgets # interactive display
plt.style.use(
"https://raw.githubusercontent.com/ClimateMatchAcademy/course-content/main/cma.mplstyle"
)
%matplotlib inline
# @title Video 1: Future Climate Scenarios
from ipywidgets import widgets
from IPython.display import YouTubeVideo
from IPython.display import IFrame
from IPython.display import display
class PlayVideo(IFrame):
def __init__(self, id, source, page=1, width=400, height=300, **kwargs):
self.id = id
if source == 'Bilibili':
src = f'https://player.bilibili.com/player.html?bvid={id}&page={page}'
elif source == 'Osf':
src = f'https://mfr.ca-1.osf.io/render?url=https://osf.io/download/{id}/?direct%26mode=render'
super(PlayVideo, self).__init__(src, width, height, **kwargs)
def display_videos(video_ids, W=400, H=300, fs=1):
tab_contents = []
for i, video_id in enumerate(video_ids):
out = widgets.Output()
with out:
if video_ids[i][0] == 'Youtube':
video = YouTubeVideo(id=video_ids[i][1], width=W,
height=H, fs=fs, rel=0)
print(f'Video available at https://youtube.com/watch?v={video.id}')
else:
video = PlayVideo(id=video_ids[i][1], source=video_ids[i][0], width=W,
height=H, fs=fs, autoplay=False)
if video_ids[i][0] == 'Bilibili':
print(f'Video available at https://www.bilibili.com/video/{video.id}')
elif video_ids[i][0] == 'Osf':
print(f'Video available at https://osf.io/{video.id}')
display(video)
tab_contents.append(out)
return tab_contents
video_ids = [('Youtube', 'XJppKGY0w0I'), ('Bilibili', 'BV1Jk4y1P7xf')]
tab_contents = display_videos(video_ids, W=730, H=410)
tabs = widgets.Tab()
tabs.children = tab_contents
for i in range(len(tab_contents)):
tabs.set_title(i, video_ids[i][0])
display(tabs)
Section 1: Load CMIP6 SST Data from Several Experiments Using xarray#
In the last tutorial we loaded data from the SSP5-8.5 (high-emissions projection) experiment of one CMIP6 model called TaiESM1.
Let’s expand on this by using data from three experiments
historical: a simulation of 1850-2015 using observed forcing,
SSP1-2.6: a future, low-emissions scenario, and
SSP5-8.5: a future, high-emissions scenario.
Due to the uncertainty in how future emissions of greenhouse gases and aerosols will change, it is useful to use a range of different emission scenarios as inputs, or forcings to climate models. These scenarios (SSPs) are datasets which are based on different socio-economic conditions in the future, such as population growth, energy consumption, energy sources, and climate policies. Learn more about forcing scenarios here, and learn more about the CMIP6 scenarios here.
To learn more about CMIP, including the different experiments/scenarios, please see our CMIP Resource Bank and the CMIP website.
# open an intake catalog containing the Pangeo CMIP cloud data
col = intake.open_esm_datastore(
"https://storage.googleapis.com/cmip6/pangeo-cmip6.json"
)
# pick the experiments you require
experiment_ids = ["historical", "ssp126", "ssp585"]
# from the full `col` object, create a subset using facet search
cat = col.search(
source_id="TaiESM1",
variable_id="tos",
member_id="r1i1p1f1",
table_id="Omon",
grid_label="gn",
experiment_id=experiment_ids,
require_all_on=[
"source_id"
], # make sure that we only get models which have all of the above experiments
)
# convert the sub-catalog into a datatree object, by opening each dataset into an xarray.Dataset (without loading the data)
kwargs = dict(
preprocess=combined_preprocessing, # apply xMIP fixes to each dataset
xarray_open_kwargs=dict(
use_cftime=True
), # ensure all datasets use the same time index
storage_options={
"token": "anon"
}, # anonymous/public authentication to google cloud storage
)
cat.esmcat.aggregation_control.groupby_attrs = ["source_id", "experiment_id"]
dt = cat.to_datatree(**kwargs)
---------------------------------------------------------------------------
KeyboardInterrupt Traceback (most recent call last)
Cell In[6], line 2
1 # open an intake catalog containing the Pangeo CMIP cloud data
----> 2 col = intake.open_esm_datastore(
3 "https://storage.googleapis.com/cmip6/pangeo-cmip6.json"
4 )
6 # pick the experiments you require
7 experiment_ids = ["historical", "ssp126", "ssp585"]
File ~/miniconda3/envs/climatematch/lib/python3.10/site-packages/intake_esm/core.py:107, in esm_datastore.__init__(self, obj, progressbar, sep, registry, read_csv_kwargs, columns_with_iterables, storage_options, **intake_kwargs)
105 self.esmcat = ESMCatalogModel.from_dict(obj)
106 else:
--> 107 self.esmcat = ESMCatalogModel.load(
108 obj, storage_options=self.storage_options, read_csv_kwargs=read_csv_kwargs
109 )
111 self.derivedcat = registry or default_registry
112 self._entries = {}
File ~/miniconda3/envs/climatematch/lib/python3.10/site-packages/intake_esm/cat.py:264, in ESMCatalogModel.load(cls, json_file, storage_options, read_csv_kwargs)
262 csv_path = f'{os.path.dirname(_mapper.root)}/{cat.catalog_file}'
263 cat.catalog_file = csv_path
--> 264 df = pd.read_csv(
265 cat.catalog_file,
266 storage_options=storage_options,
267 **read_csv_kwargs,
268 )
269 else:
270 df = pd.DataFrame(cat.catalog_dict)
File ~/miniconda3/envs/climatematch/lib/python3.10/site-packages/pandas/io/parsers/readers.py:912, in read_csv(filepath_or_buffer, sep, delimiter, header, names, index_col, usecols, dtype, engine, converters, true_values, false_values, skipinitialspace, skiprows, skipfooter, nrows, na_values, keep_default_na, na_filter, verbose, skip_blank_lines, parse_dates, infer_datetime_format, keep_date_col, date_parser, date_format, dayfirst, cache_dates, iterator, chunksize, compression, thousands, decimal, lineterminator, quotechar, quoting, doublequote, escapechar, comment, encoding, encoding_errors, dialect, on_bad_lines, delim_whitespace, low_memory, memory_map, float_precision, storage_options, dtype_backend)
899 kwds_defaults = _refine_defaults_read(
900 dialect,
901 delimiter,
(...)
908 dtype_backend=dtype_backend,
909 )
910 kwds.update(kwds_defaults)
--> 912 return _read(filepath_or_buffer, kwds)
File ~/miniconda3/envs/climatematch/lib/python3.10/site-packages/pandas/io/parsers/readers.py:577, in _read(filepath_or_buffer, kwds)
574 _validate_names(kwds.get("names", None))
576 # Create the parser.
--> 577 parser = TextFileReader(filepath_or_buffer, **kwds)
579 if chunksize or iterator:
580 return parser
File ~/miniconda3/envs/climatematch/lib/python3.10/site-packages/pandas/io/parsers/readers.py:1407, in TextFileReader.__init__(self, f, engine, **kwds)
1404 self.options["has_index_names"] = kwds["has_index_names"]
1406 self.handles: IOHandles | None = None
-> 1407 self._engine = self._make_engine(f, self.engine)
File ~/miniconda3/envs/climatematch/lib/python3.10/site-packages/pandas/io/parsers/readers.py:1661, in TextFileReader._make_engine(self, f, engine)
1659 if "b" not in mode:
1660 mode += "b"
-> 1661 self.handles = get_handle(
1662 f,
1663 mode,
1664 encoding=self.options.get("encoding", None),
1665 compression=self.options.get("compression", None),
1666 memory_map=self.options.get("memory_map", False),
1667 is_text=is_text,
1668 errors=self.options.get("encoding_errors", "strict"),
1669 storage_options=self.options.get("storage_options", None),
1670 )
1671 assert self.handles is not None
1672 f = self.handles.handle
File ~/miniconda3/envs/climatematch/lib/python3.10/site-packages/pandas/io/common.py:716, in get_handle(path_or_buf, mode, encoding, compression, memory_map, is_text, errors, storage_options)
713 codecs.lookup_error(errors)
715 # open URLs
--> 716 ioargs = _get_filepath_or_buffer(
717 path_or_buf,
718 encoding=encoding,
719 compression=compression,
720 mode=mode,
721 storage_options=storage_options,
722 )
724 handle = ioargs.filepath_or_buffer
725 handles: list[BaseBuffer]
File ~/miniconda3/envs/climatematch/lib/python3.10/site-packages/pandas/io/common.py:373, in _get_filepath_or_buffer(filepath_or_buffer, encoding, compression, mode, storage_options)
370 if content_encoding == "gzip":
371 # Override compression based on Content-Encoding header
372 compression = {"method": "gzip"}
--> 373 reader = BytesIO(req.read())
374 return IOArgs(
375 filepath_or_buffer=reader,
376 encoding=encoding,
(...)
379 mode=fsspec_mode,
380 )
382 if is_fsspec_url(filepath_or_buffer):
File ~/miniconda3/envs/climatematch/lib/python3.10/http/client.py:482, in HTTPResponse.read(self, amt)
480 else:
481 try:
--> 482 s = self._safe_read(self.length)
483 except IncompleteRead:
484 self._close_conn()
File ~/miniconda3/envs/climatematch/lib/python3.10/http/client.py:631, in HTTPResponse._safe_read(self, amt)
624 def _safe_read(self, amt):
625 """Read the number of bytes requested.
626
627 This function should be used when <amt> bytes "should" be present for
628 reading. If the bytes are truly not available (due to EOF), then the
629 IncompleteRead exception can be used to detect the problem.
630 """
--> 631 data = self.fp.read(amt)
632 if len(data) < amt:
633 raise IncompleteRead(data, amt-len(data))
File ~/miniconda3/envs/climatematch/lib/python3.10/socket.py:705, in SocketIO.readinto(self, b)
703 while True:
704 try:
--> 705 return self._sock.recv_into(b)
706 except timeout:
707 self._timeout_occurred = True
File ~/miniconda3/envs/climatematch/lib/python3.10/ssl.py:1274, in SSLSocket.recv_into(self, buffer, nbytes, flags)
1270 if flags != 0:
1271 raise ValueError(
1272 "non-zero flags not allowed in calls to recv_into() on %s" %
1273 self.__class__)
-> 1274 return self.read(nbytes, buffer)
1275 else:
1276 return super().recv_into(buffer, nbytes, flags)
File ~/miniconda3/envs/climatematch/lib/python3.10/ssl.py:1130, in SSLSocket.read(self, len, buffer)
1128 try:
1129 if buffer is not None:
-> 1130 return self._sslobj.read(len, buffer)
1131 else:
1132 return self._sslobj.read(len)
KeyboardInterrupt:
Coding Exercise 1.1#
In this tutorial and the following tutorials we will be looking at the global mean sea surface temperature. To calculate this global mean, we need to know the horizontal area of every ocean grid cell in all the models we are using.
Write code to load this ocean-grid area data using the previously shown method for SST data, noting that:
We now need a variable called areacello (area of cells in the ocean)
This variable is stored in table_id Ofx (it is from the ocean model and is fixed/constant in time)
A model’s grid does not change between experiments so you only need to get grid data from the historical experiment for each model
cat_area = col.search(
source_id="TaiESM1",
# Add the appropriate variable_id
variable_id=...,
member_id="r1i1p1f1",
# Add the appropriate table_id
table_id=...,
grid_label="gn",
# Add the appropriate experiment_id
experiment_id=[...],
require_all_on=["source_id"],
)
cat_area.esmcat.aggregation_control.groupby_attrs = ["source_id", "experiment_id"]
dt_area = cat_area.to_datatree(**kwargs)
dt_with_area = DataTree()
for model, subtree in dt.items():
metric = dt_area[model]["historical"].ds["areacello"]
dt_with_area[model] = subtree.map_over_subtree(_parse_metric, metric)
Section 2: Global Mean Sea Surface Temperature (GMSST)#
The data files above contain spatial maps of the sea surface temperature for every month of each experiment’s time period. For the rest of today’s tutorials, we’re going to focus on the global mean sea surface temperature, rather than maps, as a way to visualize the ocean’s changing temperature at a global scale\(^*\).
The global mean of a property can be calculated by integrating that variable over the surface area of Earth covered by the system (ocean, atmosphere etc.) and dividing by the total surface area of that system. For Sea Surface Temperature, \(SST(x,y)\), the global mean (\(GMSST\)) can be written as an integral over the surface of the ocean (\(S_{ocean}\)):
where \(x\) and \(y\) are horizontal coordinates (i.e. longitude and latitude). This formulation works if \(SST(x,y)\) is a spatially-continuous function, but in a global model we only know the SST of discrete grid cells rather than a continuous SST field. Integrals are only defined for continuous variables, we must instead use a summation over the grid cells (summation is the discrete equivalent of integration):
where \((i,j)\) represent the indices of the 2D spatial SST data from a CMIP6 model, and \(A\) denotes the area of each ocean grid cell, which can vary between cells/locations, as you saw in the last tutorial where TaiESM1 had irregularly-gridded output. This calculation is essentially a weighted mean of the SST across the model cells, where the weighting accounts for the varying area of cells - that is, larger cells should contribute more the global mean than smaller cells.
\(^*\)Note: we could alternatively look at ocean heat content, which depends on temperature at all depths, but it is a more intensive computation that would take too long to calculate in these tutorials.
Coding Exercise 2.1#
Complete the following code so that it calculates and plots a timeseries of global mean sea surface temperature from the TaiESM1 model for both the historical experiment and the two future projection experiments, SSP1-2.6 (low emissions) and SSP5-8.5 (high emissions).
As you complete this exercise this, consider the following questions:
In the first function, what
xarrayoperation is the following line doing, and why is it neccessary?
return ds.weighted(ds.areacello.fillna(0)).mean(['x', 'y'], keep_attrs=True)
How would your time series plot might change if you instead used took a simple mean of all the sea surface temperatures across all grid cells? (Perhaps your previous maps could provide some help here)
def global_mean(ds: xr.Dataset) -> xr.Dataset:
"""Global average, weighted by the cell area"""
return ds.weighted(ds.areacello.fillna(0)).mean(["x", "y"], keep_attrs=True)
# average every dataset in the tree globally
dt_gm = ...
fig, ax = plt.subplots()
for experiment in ["historical", "ssp126", "ssp585"]:
da = ...
...
ax.set_title("Global Mean SST from TaiESM1")
ax.set_ylabel("Global Mean SST [$^\circ$C]")
ax.set_xlabel("Year")
ax.legend()
Question 1.1: Climate Connection#
Is this plot what you expected? If so, explain what you expected, and why, from the historical experiment, and the SSP1-2.6 and SSP5-8.5 scenarios (see below for a potentially useful figure).
For context, here is Figure TS.4 from the Technical Summary of the IPCC Sixth Assessment Report, which shows how several elements of forcing differ between experiments (including historical and SSP experiments). In the video above we saw the \(CO_2\) panel of this figure:
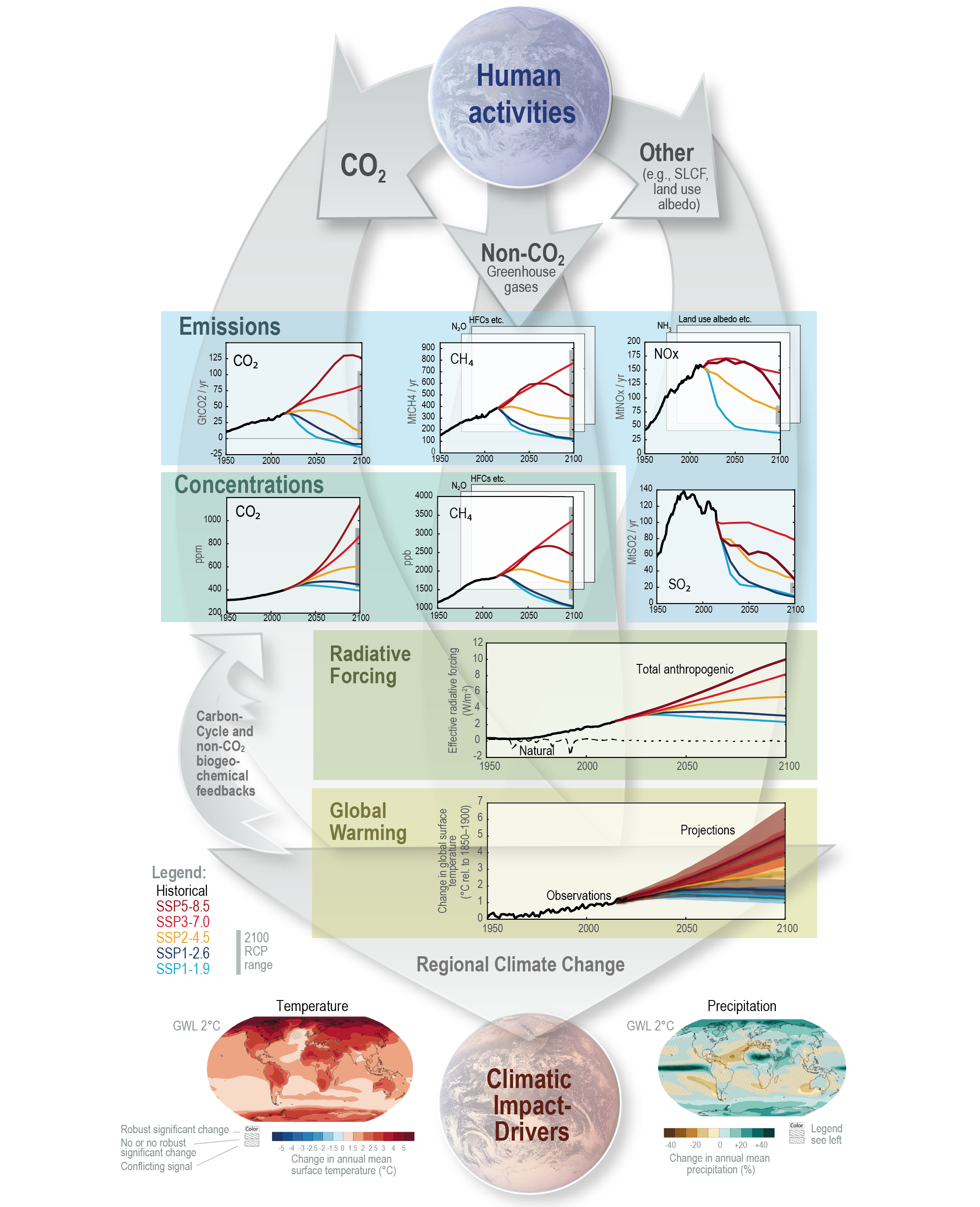
Figure TS.4 | The climate change cause–effect chain: The intent of this figure is to illustrate the process chain starting from anthropogenic emissions, to changes in atmospheric concentration, to changes in Earth’s energy balance (‘forcing’), to changes in global climate and ultimately regional climate and climatic impact-drivers. Shown is the core set of five Shared Socio-economic Pathway (SSP) scenarios as well as emissions and concentration ranges for the previous Representative Concentration Pathway (RCP) scenarios in year 2100; carbon dioxide (CO2) emissions (GtCO2yr–1), panel top left; methane (CH4) emissions (middle) and sulphur dioxide (SO2), nitrogen oxide (NOx) emissions (all in Mt yr–1), top right; concentrations of atmospheric CO2(ppm) and CH4 (ppb), second row left and right; effective radiative forcing for both anthropogenic and natural forcings (W m–2), third row; changes in global surface air temperature (°C) relative to 1850–1900, fourth row; maps of projected temperature change (°C) (left) and changes in annual-mean precipitation (%) (right) at a global warming level (GWL) of 2°C relative to 1850–1900 (see also Figure TS.5), bottom row. Carbon cycle and non-CO2 biogeochemical feedbacks will also influence the ultimate response to anthropogenic emissions (arrows on the left). {1.6.1, Cross-Chapter Box 1.4, 4.2.2, 4.3.1, 4.6.1, 4.6.2}
Credit: IPCC
Summary#
In tutorial 2, you diagnosed changes at a global scale by calculating global mean timeseries with CMIP6 model mapped data. You then synthesized and compared global mean SST evolution in various CMIP6 experiments, spanning Earth’s recent past and several future scenarios.
We started by loading CMIP6 SST data from three different scenarios: historical, SSP1-2.6 (low-emissions future), and SSP5-8.5 (high-emissions future). This process expanded our understanding of model outputs. We then focused on calculating global mean SST, by taking into account the spatially-discrete and irregularly-gridded nature of this model’s grid cells through a weighted mean. This weighted mean approach yielded the global mean SST, providing a holistic view of the Earth’s changing sea surface temperatures under multiple future climate scenarios.
Resources#
This tutorial uses data from the simulations conducted as part of the CMIP6 multi-model ensemble.
For examples on how to access and analyze data, please visit the Pangeo Cloud CMIP6 Gallery
For more information on what CMIP is and how to access the data, please see this page.


