Ocean Acidification
Contents
Ocean Acidification#
Content creators: C. Gabriela Mayorga Adame, Lidia Krinova
Content reviewers: Jenna Pearson, Abigail Bodner, Ohad Zivan, Chi Zhang
Content editors: Zane Mitrevica, Natalie Steinemann, Ohad Zivan, Chi Zhang, Jenna Pearson
Production editors: Wesley Banfield, Jenna Pearson, Chi Zhang, Ohad Zivan
Our 2023 Sponsors: NASA TOPS, Google DeepMind
# @title Project Background
from ipywidgets import widgets
from IPython.display import YouTubeVideo
from IPython.display import IFrame
from IPython.display import display
class PlayVideo(IFrame):
def __init__(self, id, source, page=1, width=400, height=300, **kwargs):
self.id = id
if source == "Bilibili":
src = f"https://player.bilibili.com/player.html?bvid={id}&page={page}"
elif source == "Osf":
src = f"https://mfr.ca-1.osf.io/render?url=https://osf.io/download/{id}/?direct%26mode=render"
super(PlayVideo, self).__init__(src, width, height, **kwargs)
def display_videos(video_ids, W=400, H=300, fs=1):
tab_contents = []
for i, video_id in enumerate(video_ids):
out = widgets.Output()
with out:
if video_ids[i][0] == "Youtube":
video = YouTubeVideo(
id=video_ids[i][1], width=W, height=H, fs=fs, rel=0
)
print(f"Video available at https://youtube.com/watch?v={video.id}")
else:
video = PlayVideo(
id=video_ids[i][1],
source=video_ids[i][0],
width=W,
height=H,
fs=fs,
autoplay=False,
)
if video_ids[i][0] == "Bilibili":
print(
f"Video available at https://www.bilibili.com/video/{video.id}"
)
elif video_ids[i][0] == "Osf":
print(f"Video available at https://osf.io/{video.id}")
display(video)
tab_contents.append(out)
return tab_contents
video_ids = [('Youtube', 'NAgrB8HxMMk'), ('Bilibili', 'BV1fM4y1x7g8')]
tab_contents = display_videos(video_ids, W=730, H=410)
tabs = widgets.Tab()
tabs.children = tab_contents
for i in range(len(tab_contents)):
tabs.set_title(i, video_ids[i][0])
display(tabs)
# @title Tutorial slides
# @markdown These are the slides for the videos in all tutorials today
from IPython.display import IFrame
link_id = "n7wdy"
Human activities release CO2 into the atmosphere, which leads to atmospheric warming and climate change. A portion of this CO2 released by human activities is absorbed into the oceans, which has a direct, chemical effect on seawater, known as ocean acidification. When CO2 combines with water in the ocean it forms carbonic acid, which makes the ocean more acidic and can have negative impacts on certain marine ecosystems (e.g., reduce the ability of calcifying organisms to form their shells and skeletons). The degree of ocean acidification is often expressed in terms of the pH of seawater, which is the measure of acidity or alkalinity such that a pH below 7 is considered acidic, and a pH greater than 7 is considered alkaline, or basic. Additional background information on ocean acidification can be found here. In this project, you will explore spatial and temporal patterns of and relationships between pH, CO2, and temperature to assess changes in ocean acidification and the impact on marine ecosystems.
In this project, you will analyse ocean model and observational data from global databases to extract variables like pH, CO2, and temperature, and to investigate ocean acidification process in your region of interest. This project will also be an opportunity to investigate the relationships between these variables as well as their impact on the marine ecosystems.
Data Exploration Notebook#
Project Setup#
# google colab installs
# !mamaba install netCDF4
# imports
import random
import numpy as np
import matplotlib.pyplot as plt
import xarray as xr
import pooch
import pandas as pd
import os
import tempfile
# helper functions
def pooch_load(filelocation=None,filename=None,processor=None):
shared_location='/home/jovyan/shared/Data/Projects/Ocean_Acidification' # this is different for each day
user_temp_cache=tempfile.gettempdir()
if os.path.exists(os.path.join(shared_location,filename)):
file = os.path.join(shared_location,filename)
else:
file = pooch.retrieve(filelocation,known_hash=None,fname=os.path.join(user_temp_cache,filename),processor=processor)
return file
NOAA Ocean pH and Acidity#
Global surface ocean acidification indicators from 1750 to 2100 (NCEI Accession 0259391)#
This data package contains a hybrid surface ocean acidification (OA) data product that is produced based on three recent observational data products:
Surface Ocean CO2 Atlas (SOCAT, version 2022)
Global Ocean Data Analysis Product version 2 (GLODAPv2, version 2022)
Coastal Ocean Data Analysis Product in North America (CODAP-NA, version 2021), and 14 Earth System Models from the sixth phase of the Coupled Model Intercomparison Project (CMIP6).
The trajectories of ten OA indicators are included in this data product:
Fugacity of carbon dioxide
pH on Total Scale
Total hydrogen ion content
Free hydrogen ion content
Carbonate ion content
Aragonite saturation state
Calcite saturation state
Revelle Factor
Total dissolved inorganic carbon content
Total alkalinity content
These OA trajectories are provided under preindustrial conditions, historical conditions, and future Shared Socioeconomic Pathways: SSP1-1.9, SSP1-2.6, SSP2-4.5, SSP3-7.0, and SSP5-8.5 from 1750 to 2100 on a global surface ocean grid. These OA trajectories are improved relative to previous OA data products with respect to data quantity, spatial and temporal coverage, diversity of the underlying data and model simulations, and the provided SSPs over the 21st century.
Citation: Jiang, L.-Q., Dunne, J., Carter, B. R., Tjiputra, J. F., Terhaar, J., Sharp, J. D., et al. (2023). Global surface ocean acidification indicators from 1750 to 2100. Journal of Advances in Modeling Earth Systems, 15, e2022MS003563. https://doi.org/10.1029/2022MS003563
Dataset: https://www.ncei.noaa.gov/data/oceans/ncei/ocads/metadata/0259391.html
We can load and visualize the surface pH as follows:
# code to retrieve and load the data
# url_SurfacepH= 'https://www.ncei.noaa.gov/data/oceans/ncei/ocads/data/0206289/Surface_pH_1770_2100/Surface_pH_1770_2000.nc' $ old CMIP5 dataset
filename_SurfacepH='pHT_median_historical.nc'
url_SurfacepH='https://www.ncei.noaa.gov/data/oceans/ncei/ocads/data/0259391/nc/median/pHT_median_historical.nc'
ds_pH = xr.open_dataset(pooch_load(url_SurfacepH,filename_SurfacepH))
ds_pH
<xarray.Dataset>
Dimensions: (time: 18, lat: 180, lon: 360)
Coordinates:
* time (time) float64 1.75e+03 1.85e+03 1.86e+03 ... 2e+03 2.01e+03
Dimensions without coordinates: lat, lon
Data variables:
pHT (time, lat, lon) float64 ...
longitude (lat, lon) float64 ...
latitude (lat, lon) float64 ...
Attributes:
title: Global surface ocean pH on total hydrogen ion scale ...
comment: This gridded data product contains pH on total hydro...
reference: Jiang, L-Q., J. Dunne, B. R. Carter, J. Tjiputra,\n ...
Fair_use_statement: This data product is made freely available\n to th...
created_by: Li-Qing Jiang
institution: (a) Cooperative Institute for Satellite Earth System...
contact: <Liqing.Jiang@noaa.gov>
creation_date: August 14, 2022For those feeling adventurouts, there are also files of future projected changes under various scenarios (SSP1-1.9, SSP1-2.6, SSP2-4.5, SSP3-7.0, and SSP5-8.5, recall W2D1 tutorials):
pHT_median_ssp119.nc
pHT_median_ssp126.nc
pHT_median_ssp245.nc
pHT_median_ssp370.nc
pHT_median_ssp585.nc
To load them, replace the filename in the path/filename line above. These data were calculated from CMIP6 models. To learn more about CMIP please see our CMIP Resource Bank and the CMIP website.
Copernicus#
Copernicus is the Earth observation component of the European Union’s Space programme, looking at our planet and its environment to benefit all European citizens. It offers information services that draw from satellite Earth Observation and in-situ (non-space) data.
The European Commission manages the Programme. It is implemented in partnership with the Member States, the European Space Agency (ESA), the European Organisation for the Exploitation of Meteorological Satellites (EUMETSAT), the European Centre for Medium-Range Weather Forecasts (ECMWF), EU Agencies and Mercator Océan.
Vast amounts of global data from satellites and ground-based, airborne, and seaborne measurement systems provide information to help service providers, public authorities, and other international organisations improve European citizens’ quality of life and beyond. The information services provided are free and openly accessible to users.
Source: https://www.copernicus.eu/en/about-copernicus
ECMWF Atmospheric Composition Reanalysis: Carbon Dioxide (CO2)#
From this dataset we will use CO2 column-mean molar fraction from the Single-level chemical vertical integrals variables & Sea Surface Temperature from the Single-level meteorological variables (in case you need to download them direclty from the catalog).
This dataset is part of the ECMWF Atmospheric Composition Reanalysis focusing on long-lived greenhouse gases: carbon dioxide (CO2) and methane (CH4). The emissions and natural fluxes at the surface are crucial for the evolution of the long-lived greenhouse gases in the atmosphere. In this dataset the CO2 fluxes from terrestrial vegetation are modelled in order to simulate the variability across a wide range of scales from diurnal to inter-annual. The CH4 chemical loss is represented by a climatological loss rate and the emissions at the surface are taken from a range of datasets.
Reanalysis combines model data with observations from across the world into a globally complete and consistent dataset using a model of the atmosphere based on the laws of physics and chemistry. This principle, called data assimilation, is based on the method used by numerical weather prediction centres and air quality forecasting centres, where every so many hours (12 hours at ECMWF) a previous forecast is combined with newly available observations in an optimal way to produce a new best estimate of the state of the atmosphere, called analysis, from which an updated, improved forecast is issued. Reanalysis works in the same way to allow for the provision of a dataset spanning back more than a decade. Reanalysis does not have the constraint of issuing timely forecasts, so there is more time to collect observations, and when going further back in time, to allow for the ingestion of improved versions of the original observations, which all benefit the quality of the reanalysis product.
Source & further information: https://ads.atmosphere.copernicus.eu/cdsapp#!/dataset/cams-global-ghg-reanalysis-egg4-monthly?tab=overview
We can load and visualize the sea surface temperature and CO2 concentration (from NOAA Global Monitoring Laboratory):
filename_CO2= 'co2_mm_gl.csv'
url_CO2= 'https://gml.noaa.gov/webdata/ccgg/trends/co2/co2_mm_gl.csv'
ds_CO2 = pd.read_csv(pooch_load(url_CO2,filename_CO2),header=55)
ds_CO2
| year | month | decimal | average | average_unc | trend | trend_unc | |
|---|---|---|---|---|---|---|---|
| 0 | 1979 | 1 | 1979.042 | 336.56 | 0.10 | 335.92 | 0.09 |
| 1 | 1979 | 2 | 1979.125 | 337.29 | 0.09 | 336.25 | 0.09 |
| 2 | 1979 | 3 | 1979.208 | 337.88 | 0.10 | 336.51 | 0.09 |
| 3 | 1979 | 4 | 1979.292 | 338.32 | 0.11 | 336.72 | 0.09 |
| 4 | 1979 | 5 | 1979.375 | 338.26 | 0.04 | 336.71 | 0.10 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 527 | 2022 | 12 | 2022.958 | 418.72 | 0.10 | 418.12 | 0.06 |
| 528 | 2023 | 1 | 2023.042 | 419.34 | 0.10 | 418.25 | 0.06 |
| 529 | 2023 | 2 | 2023.125 | 419.70 | 0.10 | 418.27 | 0.06 |
| 530 | 2023 | 3 | 2023.208 | 420.06 | 0.10 | 418.40 | 0.06 |
| 531 | 2023 | 4 | 2023.292 | 420.54 | 0.10 | 418.72 | 0.06 |
532 rows × 7 columns
# from W1D3 tutorial 6 we have Sea Surface Temprature from 1981 to the present:
# download the monthly sea surface temperature data from NOAA Physical System
# Laboratory. The data is processed using the OISST SST Climate Data Records
# from the NOAA CDR program.
# the data downloading may take 2-3 minutes to complete.
filename_sst='sst.mon.mean.nc'
url_sst = "https://osf.io/6pgc2/download/"
ds_SST = xr.open_dataset(pooch_load(url_sst,filename_sst))
ds_SST
Downloading data from 'https://osf.io/6pgc2/download/' to file '/tmp/sst.mon.mean.nc'.
---------------------------------------------------------------------------
KeyboardInterrupt Traceback (most recent call last)
Cell In[8], line 10
7 filename_sst='sst.mon.mean.nc'
8 url_sst = "https://osf.io/6pgc2/download/"
---> 10 ds_SST = xr.open_dataset(pooch_load(url_sst,filename_sst))
11 ds_SST
Cell In[5], line 10, in pooch_load(filelocation, filename, processor)
8 file = os.path.join(shared_location,filename)
9 else:
---> 10 file = pooch.retrieve(filelocation,known_hash=None,fname=os.path.join(user_temp_cache,filename),processor=processor)
12 return file
File ~/miniconda3/envs/climatematch/lib/python3.10/site-packages/pooch/core.py:239, in retrieve(url, known_hash, fname, path, processor, downloader, progressbar)
236 if downloader is None:
237 downloader = choose_downloader(url, progressbar=progressbar)
--> 239 stream_download(url, full_path, known_hash, downloader, pooch=None)
241 if known_hash is None:
242 get_logger().info(
243 "SHA256 hash of downloaded file: %s\n"
244 "Use this value as the 'known_hash' argument of 'pooch.retrieve'"
(...)
247 file_hash(str(full_path)),
248 )
File ~/miniconda3/envs/climatematch/lib/python3.10/site-packages/pooch/core.py:803, in stream_download(url, fname, known_hash, downloader, pooch, retry_if_failed)
799 try:
800 # Stream the file to a temporary so that we can safely check its
801 # hash before overwriting the original.
802 with temporary_file(path=str(fname.parent)) as tmp:
--> 803 downloader(url, tmp, pooch)
804 hash_matches(tmp, known_hash, strict=True, source=str(fname.name))
805 shutil.move(tmp, str(fname))
File ~/miniconda3/envs/climatematch/lib/python3.10/site-packages/pooch/downloaders.py:226, in HTTPDownloader.__call__(self, url, output_file, pooch, check_only)
224 progress = self.progressbar
225 progress.total = total
--> 226 for chunk in content:
227 if chunk:
228 output_file.write(chunk)
File ~/miniconda3/envs/climatematch/lib/python3.10/site-packages/requests/models.py:816, in Response.iter_content.<locals>.generate()
814 if hasattr(self.raw, "stream"):
815 try:
--> 816 yield from self.raw.stream(chunk_size, decode_content=True)
817 except ProtocolError as e:
818 raise ChunkedEncodingError(e)
File ~/miniconda3/envs/climatematch/lib/python3.10/site-packages/urllib3/response.py:628, in HTTPResponse.stream(self, amt, decode_content)
626 else:
627 while not is_fp_closed(self._fp):
--> 628 data = self.read(amt=amt, decode_content=decode_content)
630 if data:
631 yield data
File ~/miniconda3/envs/climatematch/lib/python3.10/site-packages/urllib3/response.py:567, in HTTPResponse.read(self, amt, decode_content, cache_content)
564 fp_closed = getattr(self._fp, "closed", False)
566 with self._error_catcher():
--> 567 data = self._fp_read(amt) if not fp_closed else b""
568 if amt is None:
569 flush_decoder = True
File ~/miniconda3/envs/climatematch/lib/python3.10/site-packages/urllib3/response.py:533, in HTTPResponse._fp_read(self, amt)
530 return buffer.getvalue()
531 else:
532 # StringIO doesn't like amt=None
--> 533 return self._fp.read(amt) if amt is not None else self._fp.read()
File ~/miniconda3/envs/climatematch/lib/python3.10/http/client.py:466, in HTTPResponse.read(self, amt)
463 if self.length is not None and amt > self.length:
464 # clip the read to the "end of response"
465 amt = self.length
--> 466 s = self.fp.read(amt)
467 if not s and amt:
468 # Ideally, we would raise IncompleteRead if the content-length
469 # wasn't satisfied, but it might break compatibility.
470 self._close_conn()
File ~/miniconda3/envs/climatematch/lib/python3.10/socket.py:705, in SocketIO.readinto(self, b)
703 while True:
704 try:
--> 705 return self._sock.recv_into(b)
706 except timeout:
707 self._timeout_occurred = True
File ~/miniconda3/envs/climatematch/lib/python3.10/ssl.py:1274, in SSLSocket.recv_into(self, buffer, nbytes, flags)
1270 if flags != 0:
1271 raise ValueError(
1272 "non-zero flags not allowed in calls to recv_into() on %s" %
1273 self.__class__)
-> 1274 return self.read(nbytes, buffer)
1275 else:
1276 return super().recv_into(buffer, nbytes, flags)
File ~/miniconda3/envs/climatematch/lib/python3.10/ssl.py:1130, in SSLSocket.read(self, len, buffer)
1128 try:
1129 if buffer is not None:
-> 1130 return self._sslobj.read(len, buffer)
1131 else:
1132 return self._sslobj.read(len)
KeyboardInterrupt:
cams-global-ghg-reanalysis-egg4-monthly#
From this dataset we will use CO2 column-mean molar fraction from the Single-level chemical vertical integrals variables & Sea Surface Temperature from the Single-level meteorological variables (in case you need to download them direclty from the catalog).
This dataset is part of the ECMWF Atmospheric Composition Reanalysis focusing on long-lived greenhouse gases: carbon dioxide (CO2) and methane (CH4). The emissions and natural fluxes at the surface are crucial for the evolution of the long-lived greenhouse gases in the atmosphere. In this dataset the CO2 fluxes from terrestrial vegetation are modelled in order to simulate the variability across a wide range of scales from diurnal to inter-annual. The CH4 chemical loss is represented by a climatological loss rate and the emissions at the surface are taken from a range of datasets.
Reanalysis combines model data with observations from across the world into a globally complete and consistent dataset using a model of the atmosphere based on the laws of physics and chemistry. This principle, called data assimilation, is based on the method used by numerical weather prediction centres and air quality forecasting centres, where every so many hours (12 hours at ECMWF) a previous forecast is combined with newly available observations in an optimal way to produce a new best estimate of the state of the atmosphere, called analysis, from which an updated, improved forecast is issued. Reanalysis works in the same way to allow for the provision of a dataset spanning back more than a decade. Reanalysis does not have the constraint of issuing timely forecasts, so there is more time to collect observations, and when going further back in time, to allow for the ingestion of improved versions of the original observations, which all benefit the quality of the reanalysis product.
Source & further information: https://ads.atmosphere.copernicus.eu/cdsapp#!/dataset/cams-global-ghg-reanalysis-egg4-monthly?tab=overview
filename_CO2_CAMS= 'SSTyCO2_CAMS_Copernicus_data.nc'
url_CO2_CAMS= ''
ds_CO2_CAMS =xr.open_dataset(pooch_load(url_CO2_CAMS,filename_CO2_CAMS))
ds_CO2_CAMS
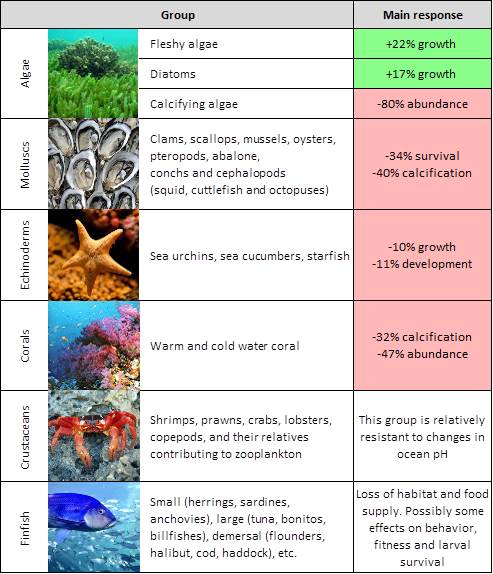
Hint for question 4:
Use the attached image (figure 5 in this website) and this mapping tool. Search for each species on the mapping tool to see the spatial global distribution.
Further Reading#
Understanding Ocean Acidification”, NOAA (https://www.fisheries.noaa.gov/insight/understanding-ocean-acidification)
“Ocean acidification and its effects”, CoastAdapt (https://coastadapt.com.au/ocean-acidification-and-its-effects)
“Scientists Pinpoint How Ocean Acidification Weakens Coral Skeletons”, WHOI (https://www.whoi.edu/press-room/news-release/scientists-identify-how-ocean-acidification-weakens-coral-skeletons/)
“Ocean acidification and reefs”, Smithonian Tropical Research Institute (https://stri.si.edu/story/ocean-acidification-and-reefs)
Henry, Joseph, Joshua Patterson, and Lisa Krimsky. 2020. “Ocean Acidification: Calcifying Marine Organisms: FA220, 3/2020”. EDIS 2020 (2):4. https://doi.org/10.32473/edis-fa220-2020.
Resources#
This tutorial uses data from the simulations conducted as part of the CMIP6 multi-model ensemble.
For examples on how to access and analyze data, please visit the Pangeo Cloud CMIP6 Gallery
For more information on what CMIP is and how to access the data, please see this page.


