Tutorial 1: Calculating ENSO with Xarray
Contents
Tutorial 1: Calculating ENSO with Xarray#
Week 1, Day 2, Ocean-Atmosphere Reanalysis
Content creators: Abigail Bodner, Momme Hell, Aurora Basinski
Content reviewers: Yosemley Bermúdez, Katrina Dobson, Danika Gupta, Maria Gonzalez, Will Gregory, Nahid Hasan, Sherry Mi, Beatriz Cosenza Muralles, Jenna Pearson, Chi Zhang, Ohad Zivan
Content editors: Jenna Pearson, Chi Zhang, Ohad Zivan
Production editors: Wesley Banfield, Jenna Pearson, Chi Zhang, Ohad Zivan
Our 2023 Sponsors: NASA TOPS and Google DeepMind
 #
#
Pythia credit: Rose, B. E. J., Kent, J., Tyle, K., Clyne, J., Banihirwe, A., Camron, D., May, R., Grover, M., Ford, R. R., Paul, K., Morley, J., Eroglu, O., Kailyn, L., & Zacharias, A. (2023). Pythia Foundations (Version v2023.05.01) https://zenodo.org/record/8065851
Tutorial Objectives#
In this notebook (adapted from Project Pythia), you will practice using multiple tools to examine sea surface temperature (SST) and explore variations in the climate system that occur during El Nino and La Nina events. You will learn to:
Load Sea Surface Temprature data from the CESM2 model
Mask data using
.where()Compute climatologies and anomalies using
.groupby()Use
.rolling()to compute moving averageCompute, normalize, and plot the Oceanic Niño Index
After completing the tasks above, you should be able to plot Oceanic Niño Index that looks similar to the figure below. The red and blue regions correspond to the phases of El Niño and La Niña respectively.
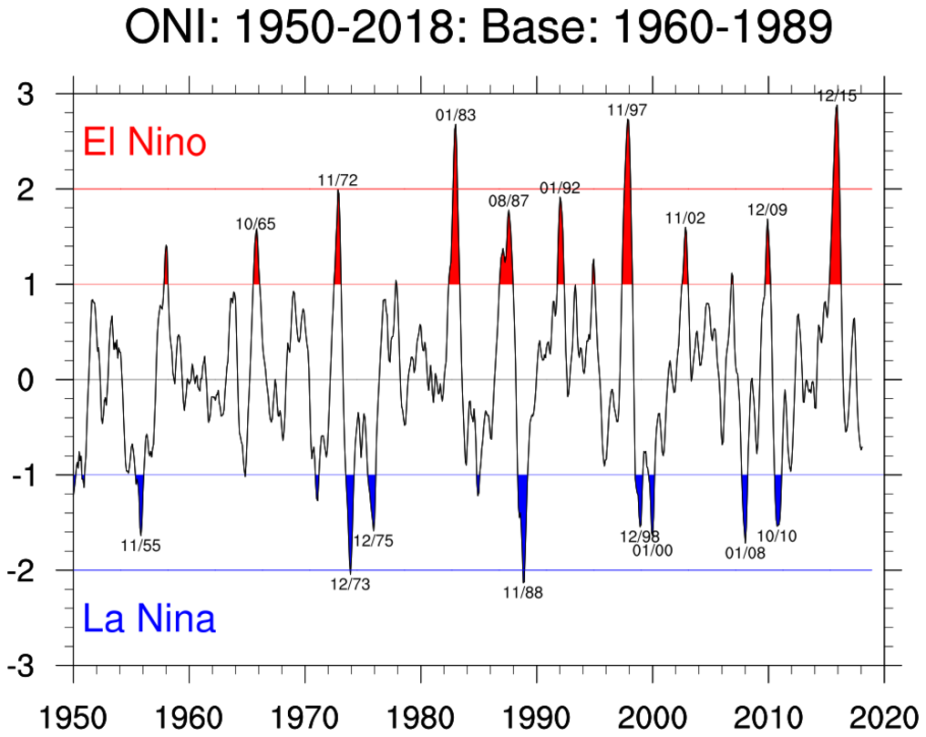
Credit: NCAR
Pythia credit: Rose, B. E. J., Kent, J., Tyle, K., Clyne, J., Banihirwe, A., Camron, D., May, R., Grover, M., Ford, R. R., Paul, K., Morley, J., Eroglu, O., Kailyn, L., & Zacharias, A. (2023). Pythia Foundations (Version v2023.05.01) https://zenodo.org/record/8065851
Setup#
# installations ( uncomment and run this cell ONLY when using google colab or kaggle )
# !pip install --upgrade --force-reinstall pythia_datasets cartopy matplotlib geoviews xarray cftime nc-time-axis
# !pip install --no-binary shapely shapely --force
# # note you will need to restart the kernel after this - there should be a small button at the end of the output.
# imports
import cartopy.crs as ccrs
import matplotlib.pyplot as plt
import xarray as xr
from pythia_datasets import DATASETS
import cartopy.io.shapereader as shapereader
import pandas as pd
import matplotlib.dates as mdates
import geoviews as gv
import geoviews.feature as gf
Section 1: Introduction to El Niño Southern Oscillation (ENSO)#
In W1D1 you practiced using Xarray to calculate a monthly climatology, climate anomalies, and a running average on monthly global Sea Surface Temperature (SST) data from the Community Earth System Model v2 (CESM2). You also used the .where() method to isolate SST data between 5ºN-5ºS and 190ºE-240ºE (or 170ºW-120ºW). This geographic region, known as the Niño 3.4 region, is in the tropical Pacific Ocean and is commonly used as a metric for determining the phase of the El Niño-Southern Oscillation (ENSO). ENSO is a recurring climate pattern involving changes in SST in the central and eastern tropical Pacific Ocean, which has two alternating phases:
El Niño: the phase of ENSO characterized by warmer than average SSTs in the central and eastern tropical Pacific Ocean, weakened east to west equatorial winds and increased rainfall in the eastern tropical Pacific.
La Niña: the phase of ENSO which is characterized by cooler than average SSTs in the central and eastern tropical Pacific Ocean, stronger east to west equatorial winds and decreased rainfall in the eastern tropical Pacific.
Section 1.1: Tropical Pacific Climate Processes#
To better understand the climate system processes that result in El Niño and La Niña events, let’s first consider typical climate conditions in the tropical Pacific Ocean. Recall from W1D1, trade winds are winds that blow east to west just north and south of the equator (these are sometimes referred to as “easterly” winds since the winds are originating from the east and blowing toward the west). And as we discussed yesterday, the reason that the trade winds blow from east to west is related to Earth’s rotation, which causes the winds in the Northern Hemisphere to curve to the right and winds in the Southern Hemisphere to curve to the left. This is known as the Coriolis effect.
If Earth’s rotation affects air movement, do you think it also influences surface ocean water movement? It does! As trade winds blow across the tropical Pacific Ocean, they move water because of friction at the ocean surface. But because of the Coriolis effect, surface water moves to the right of the wind direction in the Northern Hemisphere and to the left of the wind direction in the Southern Hemisphere. However, the speed and direction of water movement changes with depth. Ocean surface water moves at an angle to the wind, and the water under the surface water moves at a slightly larger angle, and the water below that turns at an even larger angle. The average direction of all this turning water is about a right angle from the wind direction. This average is known as Ekman transport. Since this process is driven by the trade winds, the strength of this ocean water transport varies in response to changes in the strength of the trade winds.
Section 1.2: Ocean-Atmosphere Interactions During El Niño and La Niña#
So, how does all of this relate to El Niño and La Niña? Changes in the strength of Pacific Ocean trade winds and the resulting impact on Ekman transport create variations in the tropical Pacific Ocean SST, which further results in changes to atmospheric circulation patterns and rainfall.
During an El Niño event, easterly trade winds are weaker. As a result, less warm surface water is transported to the west via Ekman transport, which causes a build-up of warm surface water in the eastern equatorial Pacific. This creates warmer than average SSTs in the eastern equatorial Pacific Ocean. The atmosphere responds to this warming with increased rising air motion and above-average rainfall in the eastern Pacific. In contrast, during a La Niña event, easterly trade winds are stronger. As a result, more warm surface water is transported to the west via Ekman transport, and cool water from deeper in the ocean rises up in the eastern Pacific during a process known as upwelling. This creates cooler than average SSTs in the eastern equatorial Pacific Ocean. This cooling decreases rising air movement in the eastern Pacific, resulting in drier than average conditions.
In this tutorial, we’ll examine SST temperatures to explore variations in the climate system that occur during El Niño and La Niña events. Specifically, we will plot and interpret CESM2 SST data from the Niño 3.4 region.
Section 2: Calculate the Oceanic Niño Index#
In this notebook, we are going to combine several topics and methods you’ve covered so far to compute the Oceanic Niño Index using SST from the CESM2 submission to the CMIP6 project.
You will be working with CMIP6 data later in the week, particularly during W2D1. You can also learn more about CMIP, including additional methods to access CMIP data, please see our CMIP Resource Bank and the CMIP website.
To calculate the Oceanic Niño Index you will:
Select SST data from Niño 3.4 region of 5ºN-5ºS and 190ºE-240ºE (or 170ºW-120ºW) shown in the figure below.
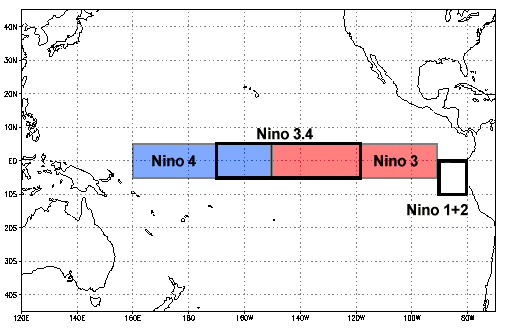
Compute the climatology (here from 2000-2014) for the Niño 3.4 region.
Compute the monthly anomaly for the Niño 3.4 region.
Compute the area-weighted mean of the anomalies for the Niño 3.4 region to obtain a time series.
Smooth the time series of anomalies with a 3-month running mean.
Here we will briefly move through each of these steps, and tomorrow you will learn about them in more detail.
Section 2.1: Open the SST Data#
First, open the SST and areacello datasets, and use Xarray’s merge method to combine them into a single dataset:
# retrive (fetch) the SST data we are going to be working on
SST_path = DATASETS.fetch("CESM2_sst_data.nc")
# open the file we acquired with xarray
SST_data = xr.open_dataset(SST_path)
# remember that one degree spatial cell is not constant around the globe, each is a different size in square km.
# we need to account for this when taking averages for example
# fetch the weight for each grid cell
gridvars_path = DATASETS.fetch("CESM2_grid_variables.nc")
# open and save only the gridcell weights whose variable name is 'areacello'
# here the 'o' at the end refers to the area cells of the 'ocean' grid
areacello_data = xr.open_dataset(gridvars_path).areacello
# merge the SST and weights into one easy to use dataset - ds stands for dataset
ds_SST = xr.merge([SST_data, areacello_data])
ds_SST
/home/wesley/miniconda3/envs/climatematch/lib/python3.10/site-packages/xarray/conventions.py:431: SerializationWarning: variable 'tos' has multiple fill values {1e+20, 1e+20}, decoding all values to NaN.
new_vars[k] = decode_cf_variable(
<xarray.Dataset>
Dimensions: (time: 180, d2: 2, lat: 180, lon: 360)
Coordinates:
* time (time) object 2000-01-15 12:00:00 ... 2014-12-15 12:00:00
* lat (lat) float64 -89.5 -88.5 -87.5 -86.5 ... 86.5 87.5 88.5 89.5
* lon (lon) float64 0.5 1.5 2.5 3.5 4.5 ... 356.5 357.5 358.5 359.5
Dimensions without coordinates: d2
Data variables:
time_bnds (time, d2) object ...
lat_bnds (lat, d2) float64 ...
lon_bnds (lon, d2) float64 ...
tos (time, lat, lon) float32 ...
areacello (lat, lon) float64 ...
Attributes: (12/45)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 674885.0
branch_time_in_parent: 219000.0
case_id: 972
... ...
sub_experiment_id: none
table_id: Omon
tracking_id: hdl:21.14100/2975ffd3-1d7b-47e3-961a-33f212ea4eb2
variable_id: tos
variant_info: CMIP6 20th century experiments (1850-2014) with C...
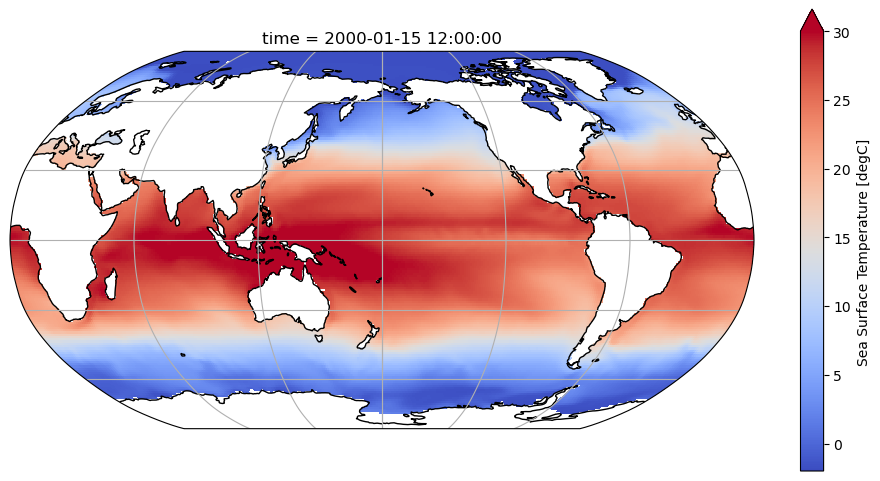
variant_label: r11i1p1f1Visualize the first time point in the early 2000s. You can check this on the indexes of the variable ‘time’ from the ds_SST above. Note that using the plot function of our dataarray will automatically include this as a title when we select just the first time index.
# define the plot size
fig = plt.figure(figsize=(12, 6))
# asssign axis and define the projection - for a round plot
ax = plt.axes(projection=ccrs.Robinson(central_longitude=180))
# add coastlines - this will issue a download warning but that is ok
ax.coastlines()
# add gridlines (lon and lat)
ax.gridlines()
# plots the first time index (0) of SST (variable name 'tos') at the first time
ds_SST.tos.isel(time=0).plot(
ax=ax,
transform=ccrs.PlateCarree(), # give our axis a map projection
vmin=-2,
vmax=30, # define the temp range of the colorbarfrom -2 to 30C
cmap="coolwarm", # choose a colormap
)
<cartopy.mpl.geocollection.GeoQuadMesh at 0x7efb83a2d120>
Interactive Demo 2.1#
You can visualize what the next few times look like in the model by using the interactive sliderbar below.
# a bit more complicated code that allows interactive plots
gv.extension("bokeh") # load Bokeh
dataset_plot = gv.Dataset(
ds_SST.isel(time=slice(0, 10))
) # slice only the first 10 timepoint, as it is a time consuming task
images = dataset_plot.to(gv.Image, ["longitude", "latitude"], "tos", "time")
images.opts(
cmap="coolwarm",
colorbar=True,
width=600,
height=400,
projection=ccrs.Robinson(),
clabel="Sea Surface Temperature [˚C]",
) * gf.coastline
Section 2.2: Select the Niño 3.4 Region#
You may have noticed that the lon for the SST data is organized between 0°–360°E.
ds_SST.lon
<xarray.DataArray 'lon' (lon: 360)>
array([ 0.5, 1.5, 2.5, ..., 357.5, 358.5, 359.5])
Coordinates:
* lon (lon) float64 0.5 1.5 2.5 3.5 4.5 ... 355.5 356.5 357.5 358.5 359.5
Attributes:
axis: X
bounds: lon_bnds
long_name: longitude
standard_name: longitude
units: degrees_eastThis is different from how we typically use longitude (-180°–180°). How do we covert the value of longitude between two systems (0-360° v.s. -180°–180°)?
Let’s use lon2 refer to the longitude system of 0°-360° while lon refers to the system of -180°–180°. 0°-360° is equivalent to 0–180°, -180°–0°.
In other words, lon2=181° is same as lon=-179°. Hence, in the western hemisphere, lon2=lon+360.
Therefore, the Niño 3.4 region should be (-5°–5°, 190–240°) using the lon2 system.
Now that we have identified the longitude values we need to select, there are a couple ways to select the Niño 3.4 region. We will demonstrate how to use both below.
sel()
# select just the Nino 3.4 region (note our longitude values are in degrees east) by slicing
tos_nino34_op1 = ds_SST.sel(lat=slice(-5, 5), lon=slice(190, 240))
tos_nino34_op1
<xarray.Dataset>
Dimensions: (time: 180, d2: 2, lat: 10, lon: 50)
Coordinates:
* time (time) object 2000-01-15 12:00:00 ... 2014-12-15 12:00:00
* lat (lat) float64 -4.5 -3.5 -2.5 -1.5 -0.5 0.5 1.5 2.5 3.5 4.5
* lon (lon) float64 190.5 191.5 192.5 193.5 ... 236.5 237.5 238.5 239.5
Dimensions without coordinates: d2
Data variables:
time_bnds (time, d2) object ...
lat_bnds (lat, d2) float64 ...
lon_bnds (lon, d2) float64 ...
tos (time, lat, lon) float32 ...
areacello (lat, lon) float64 ...
Attributes: (12/45)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 674885.0
branch_time_in_parent: 219000.0
case_id: 972
... ...
sub_experiment_id: none
table_id: Omon
tracking_id: hdl:21.14100/2975ffd3-1d7b-47e3-961a-33f212ea4eb2
variable_id: tos
variant_info: CMIP6 20th century experiments (1850-2014) with C...
variant_label: r11i1p1f1Use
where()and select all values within the bounds of interest
# select just the Nino 3.4 region (note our longitude values are in degrees east) by boolean conditioning
tos_nino34_op2 = ds_SST.where(
(ds_SST.lat < 5) & (ds_SST.lat > -5) & (ds_SST.lon > 190) & (ds_SST.lon < 240),
drop=True,
) # use dataset where function. use boolean commands
tos_nino34_op2
<xarray.Dataset>
Dimensions: (time: 180, d2: 2, lat: 10, lon: 50)
Coordinates:
* time (time) object 2000-01-15 12:00:00 ... 2014-12-15 12:00:00
* lat (lat) float64 -4.5 -3.5 -2.5 -1.5 -0.5 0.5 1.5 2.5 3.5 4.5
* lon (lon) float64 190.5 191.5 192.5 193.5 ... 236.5 237.5 238.5 239.5
Dimensions without coordinates: d2
Data variables:
time_bnds (time, d2, lat, lon) object 2000-01-01 00:00:00 ... 2015-01-01...
lat_bnds (lat, d2, lon) float64 -5.0 -5.0 -5.0 -5.0 ... 5.0 5.0 5.0 5.0
lon_bnds (lon, d2, lat) float64 190.0 190.0 190.0 ... 240.0 240.0 240.0
tos (time, lat, lon) float32 28.26 28.16 28.06 ... 28.54 28.57 28.63
areacello (lat, lon) float64 1.233e+10 1.233e+10 ... 1.233e+10 1.233e+10
Attributes: (12/45)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 674885.0
branch_time_in_parent: 219000.0
case_id: 972
... ...
sub_experiment_id: none
table_id: Omon
tracking_id: hdl:21.14100/2975ffd3-1d7b-47e3-961a-33f212ea4eb2
variable_id: tos
variant_info: CMIP6 20th century experiments (1850-2014) with C...
variant_label: r11i1p1f1You can verify that tos_nino34_op1 and tos_nino34_op2 are the same by comparing the lat and lon indexes.
We only need one of these, so let us choose the second option and set that to the variable we will use moving forward.
# SST in just the Nino 3.4 region
tos_nino34 = tos_nino34_op2
Let’s utilize the same code we used to plot the entire Earth, but this time focusing on the Niño 3.4 region slice only.
# define the figure size
fig = plt.figure(figsize=(12, 6))
# assign axis and projection
ax = plt.axes(projection=ccrs.Robinson(central_longitude=180))
# add coastlines
ax.coastlines()
# add gridlines (lon and lat)
ax.gridlines()
# plot as above
tos_nino34.tos.isel(time=0).plot(
ax=ax, transform=ccrs.PlateCarree(), vmin=-2, vmax=30, cmap="coolwarm"
)
# make sure we see more areas of the earth and not only the square around Niño 3.4
ax.set_extent((120, 300, 10, -10))
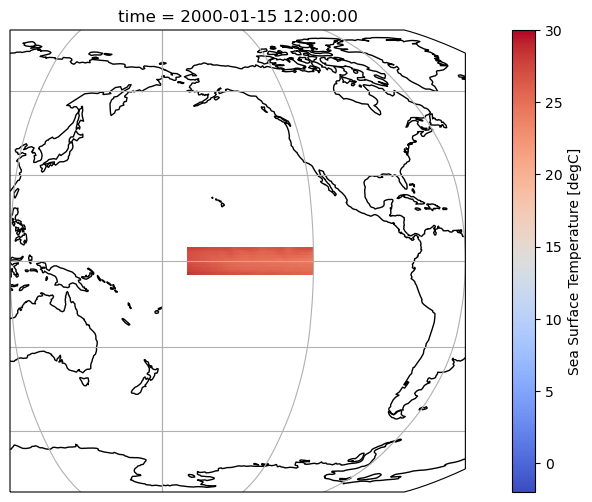
Section 2.3: Compute the Climatology and Anomalies#
Now that we have selected our area, we can compute the monthly anomaly by first grouping all the data by month, then substracting the monthly climatology from each month.
# group the dataset by month
tos_nino34_mon = tos_nino34.tos.groupby("time.month")
# find the monthly climatology in the Nino 3.4 region
tos_nino34_clim = tos_nino34_mon.mean(dim="time")
# find the monthly anomaly in the Nino 3.4 region
tos_nino34_anom = tos_nino34_mon - tos_nino34_clim
# take the area weighted average of anomalies in the Nino 3.4 region
tos_nino34_anom_mean = tos_nino34_anom.weighted(tos_nino34.areacello).mean(
dim=["lat", "lon"]
)
tos_nino34_anom_mean
<xarray.DataArray 'tos' (time: 180)>
array([-1.09020297, -0.94682973, -0.87016881, -0.67314576, -0.58964472,
-0.42163337, -0.1045906 , 0.03671601, 0.10723412, 0.17852147,
0.03699046, 0.037018 , 0.18047598, 0.13209794, 0.18591605,
0.23601163, 0.40335283, 0.41607813, 0.56452047, 0.71568734,
0.94604367, 1.54824442, 1.64511469, 1.71039068, 1.68536358,
1.59079579, 1.06137074, 0.42683044, 0.28220138, 0.01182272,
-0.50274312, -1.31489676, -1.3469001 , -1.74364307, -2.1627902 ,
-2.25649891, -2.34426791, -2.36370234, -2.04569948, -2.06573344,
-1.83853623, -1.44441816, -0.9501292 , -0.29584973, 0.02383247,
0.30428051, 0.42201282, 0.49855477, 0.48539399, 0.51069788,
0.39988395, 0.32679855, 0.06415625, -0.16908221, 0.07204665,
0.55496837, 0.65673466, 0.57821731, 0.65645898, 0.64569495,
0.83701739, 0.75305036, 0.68472177, 0.46625659, 0.27632608,
-0.16300001, -0.6672963 , -0.97958267, -1.00201317, -1.34717404,
-1.34564208, -1.56877085, -1.43570676, -0.91709921, -0.60461385,
-0.44474806, -0.39675869, -0.34406608, -0.19599503, -0.22287623,
-0.27117913, -0.64625193, -0.58092378, -0.67215087, -0.69564098,
-0.71416747, -0.51178696, -0.60251519, -0.50095793, -0.2258347 ,
-0.02674856, -0.04463503, -0.11126012, 0.14102328, 0.3094833 ,
0.28171968, 0.29325428, 0.38412098, 0.42648494, 0.75224687,
1.00070393, 1.25250361, 1.5257914 , 1.62650835, 1.6208905 ,
1.74093494, 1.82211019, 2.049507 , 2.12185874, 1.92467515,
1.41653586, 1.11950453, 0.51565086, -0.44916468, -1.64657781,
-2.17301426, -2.60500161, -2.81257123, -2.8883486 , -2.78451 ,
-2.76460858, -2.71103114, -2.78661373, -2.20402363, -2.07531219,
-1.83626328, -1.55763762, -1.53099864, -1.38395764, -1.43226824,
-1.52907282, -1.55885613, -1.05034929, -0.66684363, 0.06911014,
0.35993651, 0.32698891, 0.49531353, 0.58532671, 0.64840056,
0.83827156, 1.04578151, 1.13088586, 1.39856479, 1.74717953,
1.7443611 , 1.53452867, 1.35453215, 1.67649088, 2.10747709,
2.56691606, 2.78958853, 2.3695489 , 2.26843057, 2.38747462,
2.45266686, 2.80075567, 2.32577243, 1.74048293, 1.65214002,
1.56080466, 1.18142003, 0.47276929, 0.02291112, -0.19411324,
-0.45835808, -0.6191515 , -0.69644142, -0.77052278, -1.04589795,
-0.7001528 , -0.70409135, -0.70546689, -0.41115269, -0.13565225,
0.16707414, 0.35186853, 0.63483198, 0.71539748, 0.46310973])
Coordinates:
* time (time) object 2000-01-15 12:00:00 ... 2014-12-15 12:00:00
month (time) int64 1 2 3 4 5 6 7 8 9 10 11 ... 2 3 4 5 6 7 8 9 10 11 12Section 2.4: Smooth the Anomaly Time Series#
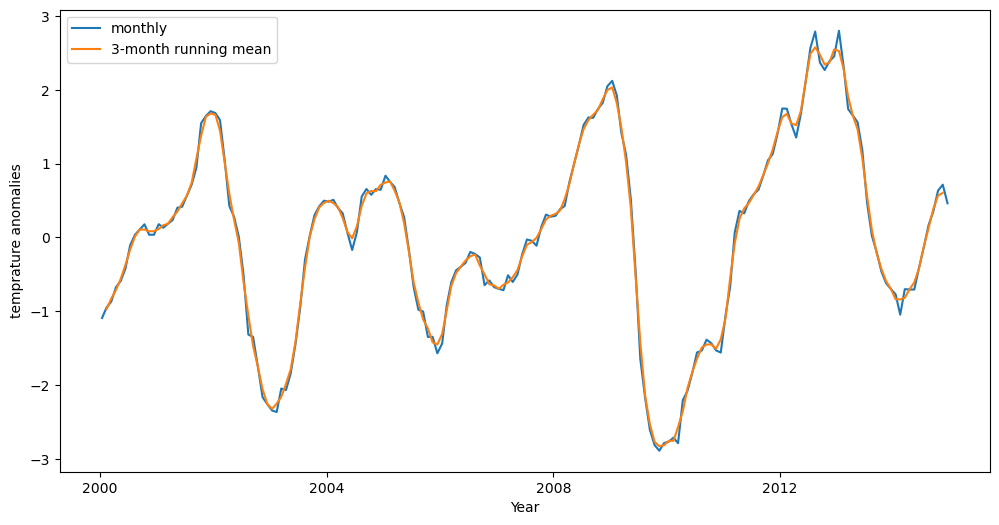
ENSO occurs on interannual timescales (a few years or more). To isolate the variability of this longer-term phenomenon on the Niño 3.4 region, we can smooth out the fluctuations due to variability on shorter timescales. To achieve this, we will apply a 3-month running mean to our time series of SST anomalies.
# smooth using a centered 3 month running mean
oni_index = tos_nino34_anom_mean.rolling(time=3, center=True).mean()
# define the plot size
fig = plt.figure(figsize=(12, 6))
# assign axis
ax = plt.axes()
# plot the monhtly data on the assigned axis
tos_nino34_anom_mean.plot(ax=ax)
# plot the smoothed data on the assigned axis
oni_index.plot(ax=ax)
# add legend
ax.legend(["monthly", "3-month running mean"])
# add ylabel
ax.set_ylabel("temprature anomalies")
# add xlabel
ax.set_xlabel("Year")
Text(0.5, 0, 'Year')
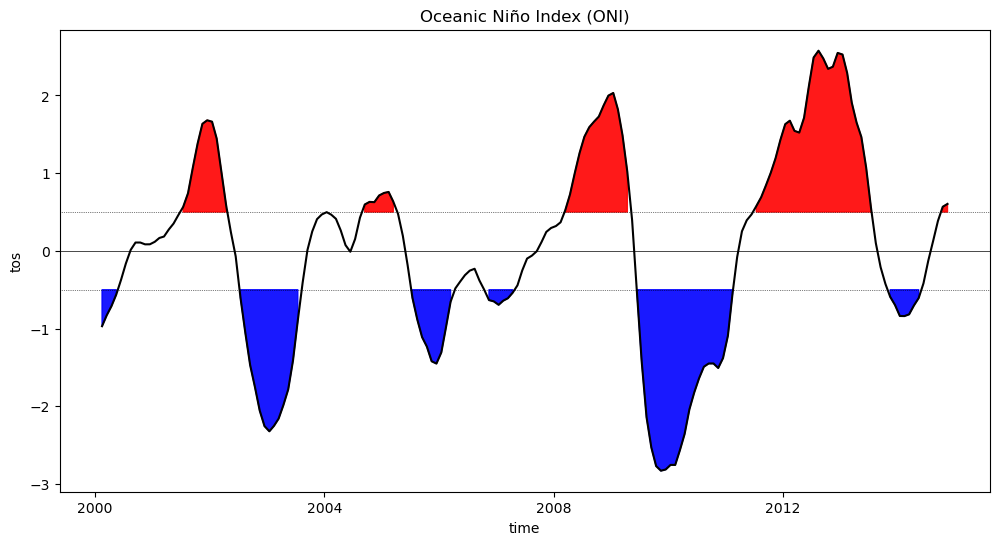
Section 3: Identify El Niño and La Niña Events#
We will highlight values in excess of \(\pm\)0.5, roughly corresponding to El Niño (warm) and La Niña (cold) events.
fig = plt.figure(figsize=(12, 6)) # assing figure size
plt.fill_between( # plot with color in between
oni_index.time.data, # x values
# top boundary - y values above 0.5
oni_index.where(oni_index >= 0.5).data,
0.5, # bottom boundary - 0.5
color="red", # color
alpha=0.9, # transparency value
)
plt.fill_between(
oni_index.time.data,
oni_index.where(oni_index <= -0.5).data,
-0.5,
color="blue",
alpha=0.9,
)
oni_index.plot(color="black") # plot the smoothed data
plt.axhline(0, color="black", lw=0.5) # add a black line at x=0
plt.axhline(
0.5, color="black", linewidth=0.5, linestyle="dotted"
) # add a black line at x=0.5
plt.axhline(
-0.5, color="black", linewidth=0.5, linestyle="dotted"
) # add a black line at x=-0.5
plt.title("Oceanic Niño Index (ONI)")
Text(0.5, 1.0, 'Oceanic Niño Index (ONI)')
Questions 3:#
Now that we’ve normalized the data and highlighted SST anomalies that correspond to El Niño (warm) and La Niña (cold) events, consider the following questions:
How frequently do El Niño and La Niña events occur over the period of time studied here?
When were the strongest El Niño and La Niña events over this time period?
Considering the ocean-atmosphere interactions that cause El Niño and La Niña events, can you hypothesize potential reasons one El Niño or La Niña event may be stronger than others?
Summary#
In this tutorial, we have learned to utilize a variety of Xarray tools to examine variations in SST during El Niño and La Niña events. We’ve practiced loading SST data from the CESM2 model, and masking data using the .where() function for focused analysis. We have also computed climatologies and anomalies using the .groupby() function, and learned to compute moving averages using the .rolling() function. Finally, we have calculated, normalized, and plotted the Oceanic Niño Index (ONI), enhancing our understanding of the El Niño Southern Oscillation (ENSO) and its impacts on global climate patterns.



