Tutorial 2: Learning Hyperparameters
Contents
Tutorial 2: Learning Hyperparameters¶
Week 1, Day 2: Linear Deep Learning
By Neuromatch Academy
Content creators: Saeed Salehi, Andrew Saxe
Content reviewers: Polina Turishcheva, Antoine De Comite, Kelson Shilling-Scrivo
Content editors: Anoop Kulkarni
Production editors: Khalid Almubarak, Gagana B, Spiros Chavlis
Tutorial Objectives¶
Training landscape
The effect of depth
Choosing a learning rate
Initialization matters
Setup¶
This a GPU-Free tutorial!
⚠ Experimental LLM-enhanced tutorial ⚠
This notebook includes Neuromatch’s experimental Chatify 🤖 functionality. The Chatify notebook extension adds support for a large language model-based “coding tutor” to the materials. The tutor provides automatically generated text to help explain any code cell in this notebook.
Note that using Chatify may cause breaking changes and/or provide incorrect or misleading information. If you wish to proceed by installing and enabling the Chatify extension, you should run the next two code blocks (hidden by default). If you do not want to use this experimental version of the Neuromatch materials, please use the stable materials instead.
To use the Chatify helper, insert the %%explain magic command at the start of any code cell and then run it (shift + enter) to access an interface for receiving LLM-based assitance. You can then select different options from the dropdown menus depending on what sort of assitance you want. To disable Chatify and run the code block as usual, simply delete the %%explain command and re-run the cell.
Note that, by default, all of Chatify’s responses are generated locally. This often takes several minutes per response. Once you click the “Submit request” button, just be patient– stuff is happening even if you can’t see it right away!
Thanks for giving Chatify a try! Love it? Hate it? Either way, we’d love to hear from you about your Chatify experience! Please consider filling out our brief survey to provide feedback and help us make Chatify more awesome!
Run the next two cells to install and configure Chatify…
%pip install -q davos
import davos
davos.config.suppress_stdout = True
Note: you may need to restart the kernel to use updated packages.
smuggle chatify # pip: git+https://github.com/ContextLab/chatify.git
%load_ext chatify
Using default configuration!
Downloading the 'cache' file.
Install and import feedback gadget¶
# @title Install and import feedback gadget
!pip3 install vibecheck datatops --quiet
from vibecheck import DatatopsContentReviewContainer
def content_review(notebook_section: str):
return DatatopsContentReviewContainer(
"", # No text prompt
notebook_section,
{
"url": "https://pmyvdlilci.execute-api.us-east-1.amazonaws.com/klab",
"name": "neuromatch_dl",
"user_key": "f379rz8y",
},
).render()
feedback_prefix = "W1D2_T2"
# Imports
import time
import numpy as np
import matplotlib
import matplotlib.pyplot as plt
Figure settings¶
# @title Figure settings
import logging
logging.getLogger('matplotlib.font_manager').disabled = True
from ipywidgets import interact, IntSlider, FloatSlider, fixed
from ipywidgets import HBox, interactive_output, ToggleButton, Layout
from mpl_toolkits.axes_grid1 import make_axes_locatable
%config InlineBackend.figure_format = 'retina'
plt.style.use("https://raw.githubusercontent.com/NeuromatchAcademy/content-creation/main/nma.mplstyle")
Plotting functions¶
# @title Plotting functions
def plot_x_y_(x_t_, y_t_, x_ev_, y_ev_, loss_log_, weight_log_):
"""
Plot train data and test results
Args:
x_t_: np.ndarray
Training dataset
y_t_: np.ndarray
Ground truth corresponding to training dataset
x_ev_: np.ndarray
Evaluation set
y_ev_: np.ndarray
ShallowNarrowNet predictions
loss_log_: list
Training loss records
weight_log_: list
Training weight records (evolution of weights)
Returns:
Nothing
"""
plt.figure(figsize=(12, 4))
plt.subplot(1, 3, 1)
plt.scatter(x_t_, y_t_, c='r', label='training data')
plt.plot(x_ev_, y_ev_, c='b', label='test results', linewidth=2)
plt.xlabel('x')
plt.ylabel('y')
plt.legend()
plt.subplot(1, 3, 2)
plt.plot(loss_log_, c='r')
plt.xlabel('epochs')
plt.ylabel('mean squared error')
plt.subplot(1, 3, 3)
plt.plot(weight_log_)
plt.xlabel('epochs')
plt.ylabel('weights')
plt.show()
def plot_vector_field(what, init_weights=None):
"""
Helper function to plot vector fields
Args:
what: string
If "all", plot vectors, trajectories and loss function
If "vectors", plot vectors
If "trajectory", plot trajectories
If "loss", plot loss function
Returns:
Nothing
"""
n_epochs=40
lr=0.15
x_pos = np.linspace(2.0, 0.5, 100, endpoint=True)
y_pos = 1. / x_pos
xx, yy = np.mgrid[-1.9:2.0:0.2, -1.9:2.0:0.2]
zz = np.empty_like(xx)
x, y = xx[:, 0], yy[0]
x_temp, y_temp = gen_samples(10, 1.0, 0.0)
cmap = matplotlib.cm.plasma
plt.figure(figsize=(8, 7))
ax = plt.gca()
if what == 'all' or what == 'vectors':
for i, a in enumerate(x):
for j, b in enumerate(y):
temp_model = ShallowNarrowLNN([a, b])
da, db = temp_model.dloss_dw(x_temp, y_temp)
zz[i, j] = temp_model.loss(temp_model.forward(x_temp), y_temp)
scale = min(40 * np.sqrt(da**2 + db**2), 50)
ax.quiver(a, b, - da, - db, scale=scale, color=cmap(np.sqrt(da**2 + db**2)))
if what == 'all' or what == 'trajectory':
if init_weights is None:
for init_weights in [[0.5, -0.5], [0.55, -0.45], [-1.8, 1.7]]:
temp_model = ShallowNarrowLNN(init_weights)
_, temp_records = temp_model.train(x_temp, y_temp, lr, n_epochs)
ax.scatter(temp_records[:, 0], temp_records[:, 1],
c=np.arange(len(temp_records)), cmap='Greys')
ax.scatter(temp_records[0, 0], temp_records[0, 1], c='blue', zorder=9)
ax.scatter(temp_records[-1, 0], temp_records[-1, 1], c='red', marker='X', s=100, zorder=9)
else:
temp_model = ShallowNarrowLNN(init_weights)
_, temp_records = temp_model.train(x_temp, y_temp, lr, n_epochs)
ax.scatter(temp_records[:, 0], temp_records[:, 1],
c=np.arange(len(temp_records)), cmap='Greys')
ax.scatter(temp_records[0, 0], temp_records[0, 1], c='blue', zorder=9)
ax.scatter(temp_records[-1, 0], temp_records[-1, 1], c='red', marker='X', s=100, zorder=9)
if what == 'all' or what == 'loss':
contplt = ax.contourf(x, y, np.log(zz+0.001), zorder=-1, cmap='coolwarm', levels=100)
divider = make_axes_locatable(ax)
cax = divider.append_axes("right", size="5%", pad=0.05)
cbar = plt.colorbar(contplt, cax=cax)
cbar.set_label('log (Loss)')
ax.set_xlabel("$w_1$")
ax.set_ylabel("$w_2$")
ax.set_xlim(-1.9, 1.9)
ax.set_ylim(-1.9, 1.9)
plt.show()
def plot_loss_landscape():
"""
Helper function to plot loss landscapes
Args:
None
Returns:
Nothing
"""
x_temp, y_temp = gen_samples(10, 1.0, 0.0)
xx, yy = np.mgrid[-1.9:2.0:0.2, -1.9:2.0:0.2]
zz = np.empty_like(xx)
x, y = xx[:, 0], yy[0]
for i, a in enumerate(x):
for j, b in enumerate(y):
temp_model = ShallowNarrowLNN([a, b])
zz[i, j] = temp_model.loss(temp_model.forward(x_temp), y_temp)
temp_model = ShallowNarrowLNN([-1.8, 1.7])
loss_rec_1, w_rec_1 = temp_model.train(x_temp, y_temp, 0.02, 240)
temp_model = ShallowNarrowLNN([1.5, -1.5])
loss_rec_2, w_rec_2 = temp_model.train(x_temp, y_temp, 0.02, 240)
plt.figure(figsize=(12, 8))
ax = plt.subplot(1, 1, 1, projection='3d')
ax.plot_surface(xx, yy, np.log(zz+0.5), cmap='coolwarm', alpha=0.5)
ax.scatter3D(w_rec_1[:, 0], w_rec_1[:, 1], np.log(loss_rec_1+0.5),
c='k', s=50, zorder=9)
ax.scatter3D(w_rec_2[:, 0], w_rec_2[:, 1], np.log(loss_rec_2+0.5),
c='k', s=50, zorder=9)
plt.axis("off")
ax.view_init(45, 260)
plt.show()
def depth_widget(depth):
"""
Simulate parameter in widget
exploring impact of depth on the training curve
(loss evolution) of a deep but narrow neural network.
Args:
depth: int
Specifies depth of network
Returns:
Nothing
"""
if depth == 0:
depth_lr_init_interplay(depth, 0.02, 0.9)
else:
depth_lr_init_interplay(depth, 0.01, 0.9)
def lr_widget(lr):
"""
Simulate parameters in widget
exploring impact of depth on the training curve
(loss evolution) of a deep but narrow neural network.
Args:
lr: float
Specifies learning rate within network
Returns:
Nothing
"""
depth_lr_init_interplay(50, lr, 0.9)
def depth_lr_interplay(depth, lr):
"""
Simulate parameters in widget
exploring impact of depth on the training curve
(loss evolution) of a deep but narrow neural network.
Args:
depth: int
Specifies depth of network
lr: float
Specifies learning rate within network
Returns:
Nothing
"""
depth_lr_init_interplay(depth, lr, 0.9)
def depth_lr_init_interplay(depth, lr, init_weights):
"""
Simulate parameters in widget
exploring impact of depth on the training curve
(loss evolution) of a deep but narrow neural network.
Args:
depth: int
Specifies depth of network
lr: float
Specifies learning rate within network
init_weights: list
Specifies initial weights of the network
Returns:
Nothing
"""
n_epochs = 600
x_train, y_train = gen_samples(100, 2.0, 0.1)
model = DeepNarrowLNN(np.full((1, depth+1), init_weights))
plt.figure(figsize=(10, 5))
plt.plot(model.train(x_train, y_train, lr, n_epochs),
linewidth=3.0, c='m')
plt.title("Training a {}-layer LNN with"
" $\eta=${} initialized with $w_i=${}".format(depth, lr, init_weights), pad=15)
plt.yscale('log')
plt.xlabel('epochs')
plt.ylabel('Log mean squared error')
plt.ylim(0.001, 1.0)
plt.show()
def plot_init_effect():
"""
Helper function to plot evolution of log mean
squared error over epochs
Args:
None
Returns:
Nothing
"""
depth = 15
n_epochs = 250
lr = 0.02
x_train, y_train = gen_samples(100, 2.0, 0.1)
plt.figure(figsize=(12, 6))
for init_w in np.arange(0.7, 1.09, 0.05):
model = DeepNarrowLNN(np.full((1, depth), init_w))
plt.plot(model.train(x_train, y_train, lr, n_epochs),
linewidth=3.0, label="initial weights {:.2f}".format(init_w))
plt.title("Training a {}-layer narrow LNN with $\eta=${}".format(depth, lr), pad=15)
plt.yscale('log')
plt.xlabel('epochs')
plt.ylabel('Log mean squared error')
plt.legend(loc='lower left', ncol=4)
plt.ylim(0.001, 1.0)
plt.show()
class InterPlay:
"""
Class specifying parameters for widget
exploring relationship between the depth
and optimal learning rate
"""
def __init__(self):
"""
Initialize parameters for InterPlay
Args:
None
Returns:
Nothing
"""
self.lr = [None]
self.depth = [None]
self.success = [None]
self.min_depth, self.max_depth = 5, 65
self.depth_list = np.arange(10, 61, 10)
self.i_depth = 0
self.min_lr, self.max_lr = 0.001, 0.105
self.n_epochs = 600
self.x_train, self.y_train = gen_samples(100, 2.0, 0.1)
self.converged = False
self.button = None
self.slider = None
def train(self, lr, update=False, init_weights=0.9):
"""
Train network associated with InterPlay
Args:
lr: float
Specifies learning rate within network
init_weights: float
Specifies initial weights of the network [default: 0.9]
update: boolean
If true, show updates on widget
Returns:
Nothing
"""
if update and self.converged and self.i_depth < len(self.depth_list):
depth = self.depth_list[self.i_depth]
self.plot(depth, lr)
self.i_depth += 1
self.lr.append(None)
self.depth.append(None)
self.success.append(None)
self.converged = False
self.slider.value = 0.005
if self.i_depth < len(self.depth_list):
self.button.value = False
self.button.description = 'Explore!'
self.button.disabled = True
self.button.button_style = 'Danger'
else:
self.button.value = False
self.button.button_style = ''
self.button.disabled = True
self.button.description = 'Done!'
time.sleep(1.0)
elif self.i_depth < len(self.depth_list):
depth = self.depth_list[self.i_depth]
# Additional assert: self.min_depth <= depth <= self.max_depth
assert self.min_lr <= lr <= self.max_lr
self.converged = False
model = DeepNarrowLNN(np.full((1, depth), init_weights))
self.losses = np.array(model.train(self.x_train, self.y_train, lr, self.n_epochs))
if np.any(self.losses < 1e-2):
success = np.argwhere(self.losses < 1e-2)[0][0]
if np.all((self.losses[success:] < 1e-2)):
self.converged = True
self.success[-1] = success
self.lr[-1] = lr
self.depth[-1] = depth
self.button.disabled = False
self.button.button_style = 'Success'
self.button.description = 'Register!'
else:
self.button.disabled = True
self.button.button_style = 'Danger'
self.button.description = 'Explore!'
else:
self.button.disabled = True
self.button.button_style = 'Danger'
self.button.description = 'Explore!'
self.plot(depth, lr)
def plot(self, depth, lr):
"""
Plot following subplots:
a. Log mean squared error vs Epochs
b. Learning time vs Depth
c. Optimal learning rate vs Depth
Args:
depth: int
Specifies depth of network
lr: float
Specifies learning rate of network
Returns:
Nothing
"""
fig = plt.figure(constrained_layout=False, figsize=(10, 8))
gs = fig.add_gridspec(2, 2)
ax1 = fig.add_subplot(gs[0, :])
ax2 = fig.add_subplot(gs[1, 0])
ax3 = fig.add_subplot(gs[1, 1])
ax1.plot(self.losses, linewidth=3.0, c='m')
ax1.set_title("Training a {}-layer LNN with"
" $\eta=${}".format(depth, lr), pad=15, fontsize=16)
ax1.set_yscale('log')
ax1.set_xlabel('epochs')
ax1.set_ylabel('Log mean squared error')
ax1.set_ylim(0.001, 1.0)
ax2.set_xlim(self.min_depth, self.max_depth)
ax2.set_ylim(-10, self.n_epochs)
ax2.set_xlabel('Depth')
ax2.set_ylabel('Learning time (Epochs)')
ax2.set_title("Learning time vs depth", fontsize=14)
ax2.scatter(np.array(self.depth), np.array(self.success), c='r')
ax3.set_xlim(self.min_depth, self.max_depth)
ax3.set_ylim(self.min_lr, self.max_lr)
ax3.set_xlabel('Depth')
ax3.set_ylabel('Optimal learning rate')
ax3.set_title("Empirically optimal $\eta$ vs depth", fontsize=14)
ax3.scatter(np.array(self.depth), np.array(self.lr), c='r')
plt.show()
Helper functions¶
# @title Helper functions
def gen_samples(n, a, sigma):
"""
Generates n samples with
`y = z * x + noise(sigma)` linear relation.
Args:
n : int
Number of datapoints within sample
a : float
Offset of x
sigma : float
Standard deviation of distribution
Returns:
x : np.array
if sigma > 0, x = random values
else, x = evenly spaced numbers over a specified interval.
y : np.array
y = z * x + noise(sigma)
"""
assert n > 0
assert sigma >= 0
if sigma > 0:
x = np.random.rand(n)
noise = np.random.normal(scale=sigma, size=(n))
y = a * x + noise
else:
x = np.linspace(0.0, 1.0, n, endpoint=True)
y = a * x
return x, y
class ShallowNarrowLNN:
"""
Shallow and narrow (one neuron per layer)
linear neural network
"""
def __init__(self, init_ws):
"""
Initialize parameters of ShallowNarrowLNN
Args:
init_ws: initial weights as a list
Returns:
Nothing
"""
assert isinstance(init_ws, list)
assert len(init_ws) == 2
self.w1 = init_ws[0]
self.w2 = init_ws[1]
def forward(self, x):
"""
The forward pass through network y = x * w1 * w2
Args:
x: np.ndarray
Input data
Returns:
y: np.ndarray
y = x * w1 * w2
"""
y = x * self.w1 * self.w2
return y
def loss(self, y_p, y_t):
"""
Mean squared error (L2)
with 1/2 for convenience
Args:
y_p: np.ndarray
Network Predictions
y_t: np.ndarray
Targets
Returns:
mse: float
Average mean squared error
"""
assert y_p.shape == y_t.shape
mse = ((y_t - y_p)**2).mean()
return mse
def dloss_dw(self, x, y_t):
"""
Partial derivative of loss with respect to weights
Args:
x : np.array
Input Dataset
y_t : np.array
Corresponding Ground Truth
Returns:
dloss_dw1: float
-mean(2 * self.w2 * x * Error)
dloss_dw2: float
-mean(2 * self.w1 * x * Error)
"""
assert x.shape == y_t.shape
Error = y_t - self.w1 * self.w2 * x
dloss_dw1 = - (2 * self.w2 * x * Error).mean()
dloss_dw2 = - (2 * self.w1 * x * Error).mean()
return dloss_dw1, dloss_dw2
def train(self, x, y_t, eta, n_ep):
"""
Gradient descent algorithm
Args:
x : np.array
Input Dataset
y_t : np.array
Corrsponding target
eta: float
Learning rate
n_ep : int
Number of epochs
Returns:
loss_records: np.ndarray
Log of loss per epoch
weight_records: np.ndarray
Log of weights per epoch
"""
assert x.shape == y_t.shape
loss_records = np.empty(n_ep) # Pre allocation of loss records
weight_records = np.empty((n_ep, 2)) # Pre allocation of weight records
for i in range(n_ep):
y_p = self.forward(x)
loss_records[i] = self.loss(y_p, y_t)
dloss_dw1, dloss_dw2 = self.dloss_dw(x, y_t)
self.w1 -= eta * dloss_dw1
self.w2 -= eta * dloss_dw2
weight_records[i] = [self.w1, self.w2]
return loss_records, weight_records
class DeepNarrowLNN:
"""
Deep but thin (one neuron per layer)
linear neural network
"""
def __init__(self, init_ws):
"""
Initialize parameters of DeepNarrowLNN
Args:
init_ws: np.ndarray
Initial weights as a numpy array
Returns:
Nothing
"""
self.n = init_ws.size
self.W = init_ws.reshape(1, -1)
def forward(self, x):
"""
Forward pass of DeepNarrowLNN
Args:
x : np.array
Input features
Returns:
y: np.array
Product of weights over input features
"""
y = np.prod(self.W) * x
return y
def loss(self, y_t, y_p):
"""
Mean squared error (L2 loss)
Args:
y_t : np.array
Targets
y_p : np.array
Network's predictions
Returns:
mse: float
Mean squared error
"""
assert y_p.shape == y_t.shape
mse = ((y_t - y_p)**2 / 2).mean()
return mse
def dloss_dw(self, x, y_t, y_p):
"""
Analytical gradient of weights
Args:
x : np.array
Input features
y_t : np.array
Targets
y_p : np.array
Network Predictions
Returns:
dW: np.ndarray
Analytical gradient of weights
"""
E = y_t - y_p # i.e., y_t - x * np.prod(self.W)
Ex = np.multiply(x, E).mean()
Wp = np.prod(self.W) / (self.W + 1e-9)
dW = - Ex * Wp
return dW
def train(self, x, y_t, eta, n_epochs):
"""
Training using gradient descent
Args:
x : np.array
Input Features
y_t : np.array
Targets
eta: float
Learning rate
n_epochs : int
Number of epochs
Returns:
loss_records: np.ndarray
Log of loss over epochs
"""
loss_records = np.empty(n_epochs)
loss_records[:] = np.nan
for i in range(n_epochs):
y_p = self.forward(x)
loss_records[i] = self.loss(y_t, y_p).mean()
dloss_dw = self.dloss_dw(x, y_t, y_p)
if np.isnan(dloss_dw).any() or np.isinf(dloss_dw).any():
return loss_records
self.W -= eta * dloss_dw
return loss_records
Set random seed¶
Executing set_seed(seed=seed) you are setting the seed
#@title Set random seed
#@markdown Executing `set_seed(seed=seed)` you are setting the seed
# For DL its critical to set the random seed so that students can have a
# baseline to compare their results to expected results.
# Read more here: https://pytorch.org/docs/stable/notes/randomness.html
# Call `set_seed` function in the exercises to ensure reproducibility.
import random
import torch
def set_seed(seed=None, seed_torch=True):
"""
Function that controls randomness. NumPy and random modules must be imported.
Args:
seed : Integer
A non-negative integer that defines the random state. Default is `None`.
seed_torch : Boolean
If `True` sets the random seed for pytorch tensors, so pytorch module
must be imported. Default is `True`.
Returns:
Nothing.
"""
if seed is None:
seed = np.random.choice(2 ** 32)
random.seed(seed)
np.random.seed(seed)
if seed_torch:
torch.manual_seed(seed)
torch.cuda.manual_seed_all(seed)
torch.cuda.manual_seed(seed)
torch.backends.cudnn.benchmark = False
torch.backends.cudnn.deterministic = True
print(f'Random seed {seed} has been set.')
# In case that `DataLoader` is used
def seed_worker(worker_id):
"""
DataLoader will reseed workers following randomness in
multi-process data loading algorithm.
Args:
worker_id: integer
ID of subprocess to seed. 0 means that
the data will be loaded in the main process
Refer: https://pytorch.org/docs/stable/data.html#data-loading-randomness for more details
Returns:
Nothing
"""
worker_seed = torch.initial_seed() % 2**32
np.random.seed(worker_seed)
random.seed(worker_seed)
Set device (GPU or CPU). Execute set_device()¶
#@title Set device (GPU or CPU). Execute `set_device()`
# especially if torch modules used.
# Inform the user if the notebook uses GPU or CPU.
def set_device():
"""
Set the device. CUDA if available, CPU otherwise
Args:
None
Returns:
Nothing
"""
device = "cuda" if torch.cuda.is_available() else "cpu"
if device != "cuda":
print("GPU is not enabled in this notebook. \n"
"If you want to enable it, in the menu under `Runtime` -> \n"
"`Hardware accelerator.` and select `GPU` from the dropdown menu")
else:
print("GPU is enabled in this notebook. \n"
"If you want to disable it, in the menu under `Runtime` -> \n"
"`Hardware accelerator.` and select `None` from the dropdown menu")
return device
SEED = 2021
set_seed(seed=SEED)
DEVICE = set_device()
Random seed 2021 has been set.
GPU is not enabled in this notebook.
If you want to enable it, in the menu under `Runtime` ->
`Hardware accelerator.` and select `GPU` from the dropdown menu
Section 1: A Shallow Narrow Linear Neural Network¶
Time estimate: ~30 mins
Video 1: Shallow Narrow Linear Net¶
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Shallow_Narrow_Linear_Net_Video")
Section 1.1: A Shallow Narrow Linear Net¶
To better understand the behavior of neural network training with gradient descent, we start with the incredibly simple case of a shallow narrow linear neural net, since state-of-the-art models are impossible to dissect and comprehend with our current mathematical tools.
The model we use has one hidden layer, with only one neuron, and two weights. We consider the squared error (or L2 loss) as the cost function. As you may have already guessed, we can visualize the model as a neural network:
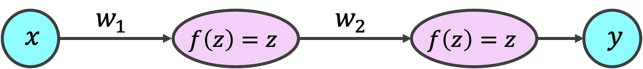
or by its computation graph:
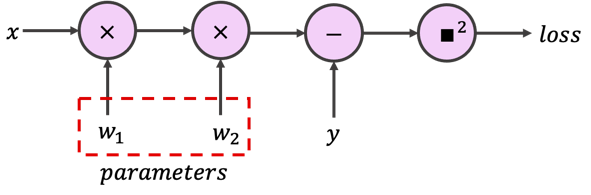
or on a rare occasion, even as a reasonably compact mapping:
Implementing a neural network from scratch without using any Automatic Differentiation tool is rarely necessary. The following two exercises are therefore Bonus (optional) exercises. Please ignore them if you have any time-limits or pressure and continue to Section 1.2.
Analytical Exercise 1.1: Loss Gradients (Optional)¶
Once again, we ask you to calculate the network gradients analytically, since you will need them for the next exercise. We understand how annoying this is.
\(\dfrac{\partial{loss}}{\partial{w_1}} = ?\)
\(\dfrac{\partial{loss}}{\partial{w_2}} = ?\)
Solution¶
\(\dfrac{\partial{loss}}{\partial{w_1}} = -2 \cdot w_2 \cdot x \cdot (y - w_1 \cdot w_2 \cdot x)\)
\(\dfrac{\partial{loss}}{\partial{w_2}} = -2 \cdot w_1 \cdot x \cdot (y - w_1 \cdot w_2 \cdot x)\)
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Loss_Gradients_Analytical_Exercise")
Coding Exercise 1.1: Implement simple narrow LNN (Optional)¶
Next, we ask you to implement the forward pass for our model from scratch without using PyTorch.
Also, although our model gets a single input feature and outputs a single prediction, we could calculate the loss and perform training for multiple samples at once. This is the common practice for neural networks, since computers are incredibly fast doing matrix (or tensor) operations on batches of data, rather than processing samples one at a time through for loops. Therefore, for the loss function, please implement the mean squared error (MSE), and adjust your analytical gradients accordingly when implementing the dloss_dw function.
Finally, complete the train function for the gradient descent algorithm:
class ShallowNarrowExercise:
"""
Shallow and narrow (one neuron per layer) linear neural network
"""
def __init__(self, init_weights):
"""
Initialize parameters of ShallowNarrow Net
Args:
init_weights: list
Initial weights
Returns:
Nothing
"""
assert isinstance(init_weights, (list, np.ndarray, tuple))
assert len(init_weights) == 2
self.w1 = init_weights[0]
self.w2 = init_weights[1]
def forward(self, x):
"""
The forward pass through netwrok y = x * w1 * w2
Args:
x: np.ndarray
Features (inputs) to neural net
Returns:
y: np.ndarray
Neural network output (predictions)
"""
#################################################
## Implement the forward pass to calculate prediction
## Note that prediction is not the loss
# Complete the function and remove or comment the line below
raise NotImplementedError("Forward Pass `forward`")
#################################################
y = ...
return y
def dloss_dw(self, x, y_true):
"""
Gradient of loss with respect to weights
Args:
x: np.ndarray
Features (inputs) to neural net
y_true: np.ndarray
True labels
Returns:
dloss_dw1: float
Mean gradient of loss with respect to w1
dloss_dw2: float
Mean gradient of loss with respect to w2
"""
assert x.shape == y_true.shape
#################################################
## Implement the gradient computation function
# Complete the function and remove or comment the line below
raise NotImplementedError("Gradient of Loss `dloss_dw`")
#################################################
dloss_dw1 = ...
dloss_dw2 = ...
return dloss_dw1, dloss_dw2
def train(self, x, y_true, lr, n_ep):
"""
Training with Gradient descent algorithm
Args:
x: np.ndarray
Features (inputs) to neural net
y_true: np.ndarray
True labels
lr: float
Learning rate
n_ep: int
Number of epochs (training iterations)
Returns:
loss_records: list
Training loss records
weight_records: list
Training weight records (evolution of weights)
"""
assert x.shape == y_true.shape
loss_records = np.empty(n_ep) # Pre allocation of loss records
weight_records = np.empty((n_ep, 2)) # Pre allocation of weight records
for i in range(n_ep):
y_prediction = self.forward(x)
loss_records[i] = loss(y_prediction, y_true)
dloss_dw1, dloss_dw2 = self.dloss_dw(x, y_true)
#################################################
## Implement the gradient descent step
# Complete the function and remove or comment the line below
raise NotImplementedError("Training loop `train`")
#################################################
self.w1 -= ...
self.w2 -= ...
weight_records[i] = [self.w1, self.w2]
return loss_records, weight_records
def loss(y_prediction, y_true):
"""
Mean squared error
Args:
y_prediction: np.ndarray
Model output (prediction)
y_true: np.ndarray
True label
Returns:
mse: np.ndarray
Mean squared error loss
"""
assert y_prediction.shape == y_true.shape
#################################################
## Implement the MEAN squared error
# Complete the function and remove or comment the line below
raise NotImplementedError("Loss function `loss`")
#################################################
mse = ...
return mse
set_seed(seed=SEED)
n_epochs = 211
learning_rate = 0.02
initial_weights = [1.4, -1.6]
x_train, y_train = gen_samples(n=73, a=2.0, sigma=0.2)
x_eval = np.linspace(0.0, 1.0, 37, endpoint=True)
## Uncomment to run
# sn_model = ShallowNarrowExercise(initial_weights)
# loss_log, weight_log = sn_model.train(x_train, y_train, learning_rate, n_epochs)
# y_eval = sn_model.forward(x_eval)
# plot_x_y_(x_train, y_train, x_eval, y_eval, loss_log, weight_log)
Random seed 2021 has been set.
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Simple_Narrow_LNN_Exercise")
Section 1.2: Learning landscapes¶
Video 2: Training Landscape¶
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Training_Landscape_Video")
As you may have already asked yourself, we can analytically find \(w_1\) and \(w_2\) without using gradient descent:
In fact, we can plot the gradients, the loss function and all the possible solutions in one figure. In this example, we use the \(y = 1x\) mapping:
Blue ribbon: shows all possible solutions: \(~ w_1 w_2 = \dfrac{y}{x} = \dfrac{x}{x} = 1 \Rightarrow w_1 = \dfrac{1}{w_2}\)
Contour background: Shows the loss values, red being higher loss
Vector field (arrows): shows the gradient vector field. The larger yellow arrows show larger gradients, which correspond to bigger steps by gradient descent.
Scatter circles: the trajectory (evolution) of weights during training for three different initializations, with blue dots marking the start of training and red crosses ( x ) marking the end of training. You can also try your own initializations (keep the initial values between -2.0 and 2.0) as shown here:
plot_vector_field('all', [1.0, -1.0])
Finally, if the plot is too crowded, feel free to pass one of the following strings as argument:
plot_vector_field('vectors') # For vector field
plot_vector_field('trajectory') # For training trajectory
plot_vector_field('loss') # For loss contour
Think!
Explore the next two plots. Try different initial values. Can you find the saddle point? Why does training slow down near the minima?
plot_vector_field('all')

Here, we also visualize the loss landscape in a 3-D plot, with two training trajectories for different initial conditions. Note: the trajectories from the 3D plot and the previous plot are independent and different.
plot_loss_landscape()

Video 3: Training Landscape - Discussion¶
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Training_Landscape_Discussion_Video")
Section 2: Depth, Learning rate, and initialization¶
Time estimate: ~45 mins
Successful deep learning models are often developed by a team of very clever people, spending many many hours “tuning” learning hyperparameters, and finding effective initializations. In this section, we look at three basic (but often not simple) hyperparameters: depth, learning rate, and initialization.
Section 2.1: The effect of depth¶
Video 4: Effect of Depth¶
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Effect_of_Depth_Video")
Why might depth be useful? What makes a network or learning system “deep”? The reality is that shallow neural nets are often incapable of learning complex functions due to data limitations. On the other hand, depth seems like magic. Depth can change the functions a network can represent, the way a network learns, and how a network generalizes to unseen data.
So let’s look at the challenges that depth poses in training a neural network. Imagine a single input, single output linear network with 50 hidden layers and only one neuron per layer (i.e. a narrow deep neural network). The output of the network is easy to calculate:
If the initial value for all the weights is \(w_i = 2\), the prediction for \(x=1\) would be exploding: \(y_p = 2^{50} \approx 1.1256 \times 10^{15}\). On the other hand, for weights initialized to \(w_i = 0.5\), the output is vanishing: \(y_p = 0.5^{50} \approx 8.88 \times 10^{-16}\). Similarly, if we recall the chain rule, as the graph gets deeper, the number of elements in the chain multiplication increases, which could lead to exploding or vanishing gradients. To avoid such numerical vulnerablities that could impair our training algorithm, we need to understand the effect of depth.
Interactive Demo 2.1: Depth widget¶
Use the widget to explore the impact of depth on the training curve (loss evolution) of a deep but narrow neural network.
Think!
Which networks trained the fastest? Did all networks eventually “work” (converge)? What is the shape of their learning trajectory?
Make sure you execute this cell to enable the widget!
# @markdown Make sure you execute this cell to enable the widget!
_ = interact(depth_widget,
depth = IntSlider(min=0, max=51,
step=5, value=0,
continuous_update=False))
Video 5: Effect of Depth - Discussion¶
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Effect_of_Depth_Discussion_Video")
Section 2.2: Choosing a learning rate¶
The learning rate is a common hyperparameter for most optimization algorithms. How should we set it? Sometimes the only option is to try all the possibilities, but sometimes knowing some key trade-offs will help guide our search for good hyperparameters.
Video 6: Learning Rate¶
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Learning_Rate_Video")
Interactive Demo 2.2: Learning rate widget¶
Here, we fix the network depth to 50 layers. Use the widget to explore the impact of learning rate \(\eta\) on the training curve (loss evolution) of a deep but narrow neural network.
Think!
Can we say that larger learning rates always lead to faster learning? Why not?
Make sure you execute this cell to enable the widget!
# @markdown Make sure you execute this cell to enable the widget!
_ = interact(lr_widget,
lr = FloatSlider(min=0.005, max=0.045, step=0.005, value=0.005,
continuous_update=False, readout_format='.3f',
description='eta'))
Video 7: Learning Rate - Discussion¶
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Learning_Rate_Discussion_Video")
Section 2.3: Depth vs Learning Rate¶
Video 8: Depth and Learning Rate¶
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Depth_and_Learning_Rate_Video")
Interactive Demo 2.3: Depth and Learning Rate¶
Important instruction The exercise starts with 10 hidden layers. Your task is to find the learning rate that delivers fast but robust convergence (learning). When you are confident about the learning rate, you can Register the optimal learning rate for the given depth. Once you press register, a deeper model is instantiated, so you can find the next optimal learning rate. The Register button turns green only when the training converges, but does not imply the fastest convergence. Finally, be patient :) the widgets are slow.
Think!
Can you explain the relationship between the depth and optimal learning rate?
Make sure you execute this cell to enable the widget!
# @markdown Make sure you execute this cell to enable the widget!
intpl_obj = InterPlay()
intpl_obj.slider = FloatSlider(min=0.005, max=0.105, step=0.005, value=0.005,
layout=Layout(width='500px'),
continuous_update=False,
readout_format='.3f',
description='eta')
intpl_obj.button = ToggleButton(value=intpl_obj.converged, description='Register')
widgets_ui = HBox([intpl_obj.slider, intpl_obj.button])
widgets_out = interactive_output(intpl_obj.train,
{'lr': intpl_obj.slider,
'update': intpl_obj.button,
'init_weights': fixed(0.9)})
display(widgets_ui, widgets_out)
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Depth_and_Learning_Rate_Interactive_Demo")
Video 9: Depth and Learning Rate - Discussion¶
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Depth_and_Learning_Rate_Discussion_Video")
Section 2.4: Why initialization is important¶
Video 10: Initialization Matters¶
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Initialization_Matters")
We’ve seen, even in the simplest of cases, that depth can slow learning. Why? From the chain rule, gradients are multiplied by the current weight at each layer, so the product can vanish or explode. Therefore, weight initialization is a fundamentally important hyperparameter.
Although in practice initial values for learnable parameters are often sampled from different \(\mathcal{Uniform}\) or \(\mathcal{Normal}\) probability distribution, here we use a single value for all the parameters.
The figure below shows the effect of initialization on the speed of learning for the deep but narrow LNN. We have excluded initializations that lead to numerical errors such as nan or inf, which are the consequence of smaller or larger initializations.
Make sure you execute this cell to see the figure!
# @markdown Make sure you execute this cell to see the figure!
plot_init_effect()

Video 11: Initialization Matters - Discussion¶
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Initialization_Matters_Discussion_Video")
Summary¶
In the second tutorial, we have learned what is the training landscape, and also we have see in depth the effect of the depth of the network and the learning rate, and their interplay. Finally, we have seen that initialization matters and why we need smart ways of initialization.
Video 12: Tutorial 2 Wrap-up¶
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_WrapUp_Video")
Bonus¶
Hyperparameter interaction¶
Finally, let’s put everything we learned together and find best initial weights and learning rate for a given depth. By now you should have learned the interactions and know how to find the optimal values quickly. If you get numerical overflow warnings, don’t be discouraged! They are often caused by “exploding” or “vanishing” gradients.
Think!
Did you experience any surprising behaviour or difficulty finding the optimal parameters?
Make sure you execute this cell to enable the widget!
# @markdown Make sure you execute this cell to enable the widget!
_ = interact(depth_lr_init_interplay,
depth = IntSlider(min=10, max=51, step=5, value=25,
continuous_update=False),
lr = FloatSlider(min=0.001, max=0.1,
step=0.005, value=0.005,
continuous_update=False,
readout_format='.3f',
description='eta'),
init_weights = FloatSlider(min=0.1, max=3.0,
step=0.1, value=0.9,
continuous_update=False,
readout_format='.3f',
description='initial weights'))
Submit your feedback¶
# @title Submit your feedback
content_review(f"{feedback_prefix}_Hyperparameter_interaction_Bonus_Discussion")

